3505
Integration of Open Source Pulse Sequence Programming Toolbox, Pulseq, for use with System Agnostic Broadband Spectrometer1Electrical and Computer Engineering, Texas A&M University, College Station, TX, United States
Synopsis
Introduced is a new translator for the open source pulse sequence programming toolbox, Pulseq. Unique from previous translator units developed, this translator takes the Pulseq output file and generates a time-bin based, delimited pulse sequence table. This format is specific to the target system, a homebuilt broadband spectrometer. Our lab uses multiple scanners, and using Pulseq enables seamless transition between a conventional research scanner, a Varian Unity Inova or a prototype homebuilt spectrometer. Ultimately, this translator is not only a useful tool, but it introduces the possibility of facilitating a direct comparison of homebuilt systems with their commercial counterparts.
Introduction
Pulse sequence development can be challenging, even for the experienced individual – the added complexity of system specific languages makes the task even more challenging. In 201_, Layton et al.1, published Pulseq, an open source toolbox that has the potential to be a powerful, platform-agnostic tool in pulse sequence writing. Presently, there are interpreter units for most of the mainstream systems, which lends value in the ability to translate sequences between them. However, we introduce a translator targeted for use on a homebuilt broadband spectrometer2. This translator not only provides a useful tool for users of the homebuilt spectrometer, but also showcases the importance of the open source initiative in aiding in reproducibility of experiments between systems. Using Pulseq with multiple translators benefits both students in NMR courses as well as researchers by providing a system agnostic programming environment.Methods
Building on previously developed Pulseq translators3, the main function of this translator is to aid in sequence programming for a homebuilt broadband spectrometer, and to provide seamless transition between a previously developed Varian Pulseq module and our homebuilt broadband spectrometer. Previously on the system, sequence writing was accomplished by either modifying a tab-delimited file by hand, or modifying an existing sequence-specific table generator. These script generators have a set sequence structure, and generate table based off timing inputs but lack flexibility to modify sequence events.The Matlab based translator module generates a tab delimited file that can then be uploaded to the spectrometer software. It is that the output file is specifically formatted for use with the homebuilt system. While previous translator units build to system specific languages, this one generates a comparatively simple delimited table. A comparison of outputs can be seen in Figure 1. This file type requires individual time bins for the trapezoidal ramp up and ramp downs, the primary challenge was finding the necessary number of time bins to accurately represent the sequence, and expand the trapezoidal waveform from a single event to three separate ones.
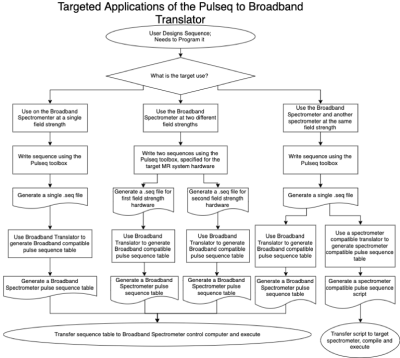
The translator is Matlab based, it could be housed on the user’s computer or the system’s control computer, as the output sequence from the translator can be easily transferred to the target system. The translator was designed such that the user would be able to write their pulse sequence using the Pulseq toolbox, generate the .seq file output file using the Pulseq toolbox, then call the broadband spectrometer’s translator to generate the pulse sequence in the compatible table format. This file can then be transferred to the broadband spectrometer’s control computer via USB, loaded into the control interface and executed normally. This process is outlined in Figure 2.
Results
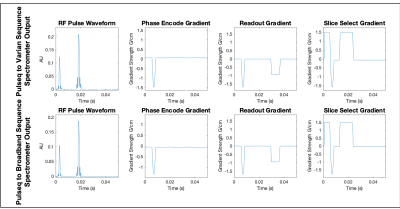
As a means to evaluate the performance of the new translator, a spin echo sequence was written in Pulseq. The .seq file was then translated using the respective system specific interpreters – one output designated for a Varian system, and one output for the homebuilt broadband spectrometer. From there, the analog outputs from the spectrometers that provide the waveforms to the RF and gradient amplifier were sampled using a digital scope. The sequences, shown in Figure 3, are observed to be comparable.For this sequence, the parameters specified were a TR time of 1000ms, a TE of 30ms, a square 100mm FOV with a 256x256 matrix, a 40kHz spectral width, for a 2mm transverse slice. RF pulses were sinc shaped, with a 4000ms duration.
Discussion
Fundamentally, this translator unit facilitates the integration of the Pulseq toolbox with a homebuilt broadband spectrometer, allowing the device to be used for both research and educational purposes. While a useful tool for those using the broadband spectrometer for research, the translator also introduces the possibility of educational applications for the broadband system. The broadband system is designed to be just that – broadband, thus it can be used at a variety of frequencies and field strengths. This would ultimately allow for students to run near identical sequences, adjusting only for the center frequencies and to avoid any hardware limitations, at different field strengths for direct comparison.The next step in the process is to fully integrate the broadband translator generated waveforms with the system amplifiers, verify that the values are correct, and begin imaging. The homebuilt broadband spectrometer is an ever-evolving project, and thus the translator for the system could naturally evolve with it. Further developments could include integrating the Pulseq toolbox such that it can be paired with an interpreter and effectively control the broadband spectrometer, generating the waveforms specifically rather than relying on shape files to be called.
Conclusion
Here we have introduced a translator that not only aids in research and educational applications, but also showcases the promising versatility of the programs and tools resulting from open-source initiatives. Also demonstrated is the broadband translator’s capability to facilitate experiment repeatability between commercial spectrometers and that of a homebuilt broadband spectrometer by producing sequences that generate comparable analog outputs.Acknowledgements
No acknowledgement found.References
1. Layton K, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen J, Stöcker T and M. Zaitsev, Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magnetic Resonance in Medicine. 2016;77(4):1544-1552.
2. Ogier, S., Bosshard, J.C., and Wright, S.M. “A Broadband Spectrometer for Simultaneous Multinuclear Magnetic Resonance Imaging and Spectroscopy.” Proc. Intl. Soc. Magn. Reson. Med., p. 546, 2016.
3. Bauer, C., and Wright, S.M. “ Development of Interpreter Module for Generating Varian VNMRJ Compatible Pulse Sequences using Pulseq Open-Source Toolbox.” Proc. Intl. Soc. Magn. Reson. Med., Poster 4840, 2019.
Figures