3503
Deep Convolutional Neural Network model for sub-Anatomy specific Landmark detection on Brain MRI1Philips Healthcare, Bangalore, India
Synopsis
A Deep CNN (D-CNN) model for Brain sub-anatomy landmark detection for auto MRI scan planning. We compare D-CNN with traditional approaches like segmentation followed by image processing (AL-Net). D-CNN shows better landmark detection with Average RMSE <= 6mm (N=100) compared to AL-Net.
Synopsis
We present a Deep Convolutional neural network (D-CNN) model for brain landmark detection, an important step in planning MRI scans for two anatomies - Corpus callosum and Cerebellum on 3D T1w images. Unlike traditional approaches like segmentation followed by image processing to identify the brain landmark points (AL-Net), we use a D-CNN to directly predict the landmarks. For all the landmarks, we demonstrate that D-CNN can achieve landmark detection with Average Root mean square error(RMSE) <= 6mm (N=100) for all landmarks and is comparable or better than AL-NET (RMSE <=8mm). Results indicate excellent feasibility of the method for clinical usage.Introduction
Landmark detection is used in several tasks, for example- facial landmark detection for facial analysis tasks 1- emotion recognition, head pose estimation, etc. MRI landmark detection plays an important role in various MRI clinical workflows and MR image analysis – scan planning 2, image registration 3 etc. There are several ways to define landmarks: 3 i. key point based (corner, edge etc.), ii. Atlas based/anatomy specific. In this work, we have focused on the anatomy specific landmark definition. For medical imaging segmentation, there are plenty of public datasets available 4,5 but there is a huge scarcity of landmark datasets in medical imaging. Also, the proposed D-CNN approach has lesser annotation effort, avoiding anatomy annotations compared to AL-Net and can be easily extended for other anatomies (for example: knee, liver etc.) and applications (like registration, etc.) in future.Materials and Methods
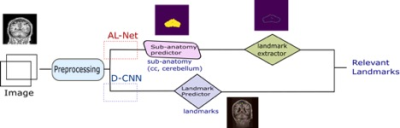
Data: For corpus callosum and cerebellum segmentation data, 2-D slices are extracted from abide-cc 4 OASIS 5 datasets. For the landmark detection dataset, first Landmark definition was created in-house (number of landmarks: 10- axial view, 7 – sagittal view, 5- coronal view) followed by in-house annotation by a Radiologist. Corpus callosum related landmarks are annotated in the sagittal view and cerebellum in the coronal view. 500 slices in each sagittal, coronal and axial view containing relevant landmarks are annotated. As shown in fig. 1, to predict the final output, there are two approaches- AL-Net and D-CNN. AL-Net is using the anatomical model output (segmentation) followed by conventional image processing approaches for landmark extraction. The D-CNN model directly predicts landmarks. The Components of fig. 1 are described below.Preprocessing: Intensity normalization, data augmentation, synthetic data synthesis are used to make the network robust to MR scanner and patient variations. Training and experimental setup: The dataset is split into three parts- 60% for training, 20% for validation and 20% for test. A Test set is used to compare both AL-Net and D-CNN.
AL-Net :
1.1. Sub-anatomy predictor (A-Net): Deep CNN network segments the anatomies in the brain region.
1.2. Landmark Extractor: It uses the output of the sub-anatomy predictor network as input and extracts landmarks using conventional image processing (detecting extreme right, extreme left, highest point etc.) for the landmark based methods. It is implemented for corpus callosum and cerebellum points only.
D-CNN:
2.1. Landmark predictor: D-CNN model uses deep CNNs for landmark detection Training: Custom deep CNN is initialized with pre-trained weights is trained separately for three different views – sagittal, coronal and axial with the input image size as (256x256) , model output as (kx64x64) where k is the number of landmarks.
Results
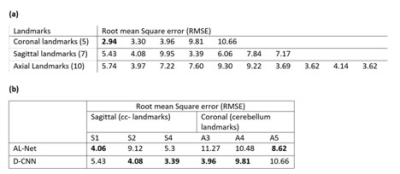
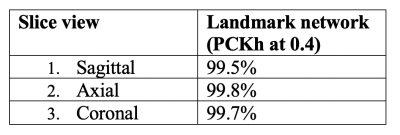
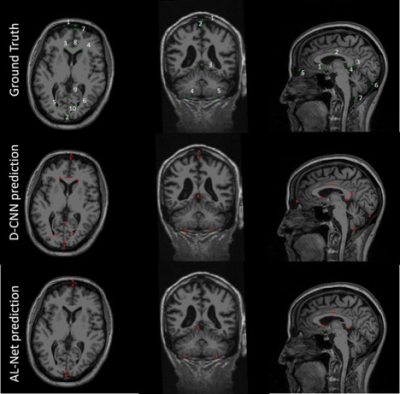
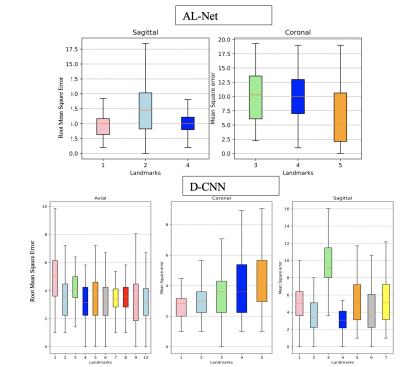
A-Net in AL-Net gives high segmentation accuracy with dice coefficient > 0.92 (slice wise) for corpus-callosum and cerebellum. Results shown in table 1 indicate that D-CNN can achieve landmark detection with Average Root mean square error(RMSE) <= 6mm (N=100) for all landmarks and is giving comparable/ better results than AL-NET (RMSE <=8mm). Also a standard metric, the PCKh (head-normalized probability of correct keypoint) score is calculated for D-CNN and is shown in Table 2, (PCKh@0.4 > 99% for all three views). Figure 2. shows the prediction of D-CNN and AL-Net compared to the ground truth. RMSE boxplot for all landmarks for D-CNN and AL-NET is shown in Figure 3. RMSE value for almost all landmarks in D-CNN is less than 10 which confirms the robustness of D-CNN model. Also accurate landmark estimation after getting A-Net segmented output is a huge task in itself, requires many image processing steps which makes it difficult to scale as number of landmarks increase.Conclusion
Result for AL-Net greatly depends on the accuracy of the segmentation model and the algorithm extracting landmarks from the corresponding segmentation. Also variation in the ground truth dataset creation for both segmentation and landmark cases is an important factor affecting the accuracies requiring more robust landmark definition. D-CNN is more generic, reduces the pipeline changes and can be extended to other anatomies and regions faster as compared to segmentation based approaches and also. In future, D-CNN output heatmaps need not be downsampled, to increase the output and number of parameters in the network, which can further improve accuracy.Acknowledgements
We would like to thank Vineeth VS, clinical specialist, PIC Bangalore for providing landmark definition for brain data annotation.References
1. Wu, Yue, and Qiang Ji. "Facial landmark detection: A literature survey." International Journal of Computer Vision 127.2 (2019): 115-142.
2. A generalized deep learning framework for multi-landmark intelligent slice placement using standard tri-planar 2D localizers , ISMRM 2019
3. Zhang, Jun, Mingxia Liu, and Dinggang Shen. Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Transactions on Image Processing 26.10 (2017): 4753-4764
4. Kucharsky Hiess, R., Alter, R.A., Sojoudi, S., Ardekani, B., Kuzniecky, R., and Pardoe, H.R. (2015) Corpus callosum area and brain volume in autism spectrum disorder: quantitative analysis of structural MRI from the ABIDE database, Journal of Autism and Developmental Disorders, 45(10): 3107-3114, doi: 10.1007/s10803-015-2468-8
5. Open Access Series of Imaging Studies (OASIS), online: http://www.oasis- brains.org
6. Sun, K., Xiao, B., Liu, D. and Wang, J., 2019. Deep high-resolution representation learning for human pose estimation. arXiv preprint arXiv:1902.09212
7. M. Andriluka, L. Pishchulin, P. V. Gehler, and B. Schiele. 2d human pose estimation: New benchmark and state of the art analysis. In CVPR, pages 3686–3693, 2014
Figures