3495
Weakly Supervised Deep Prior Learning for Multi-coil MRI Reconstruction1Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, P.R.China, Shenzhen, China
Synopsis
MRI reconstruction based on supervised learning methods, have achieved remarkable success. However, because of the difficulty and high cost of the MR images collection process, it is not always easy to obtain a big dataset with strong supervised information. Therefore, weakly supervised learning will be a possible solution. In this work, we proposed a weakly deep prior learning algorithm to train a complex UNet for multi-coil MR images reconstruction without very precise labels. The result shows that our proposed algorithm can provide a competitive performance compared to the classical methods with enoucraging quantitative indicators of SSIM and PSNR.
INTRODUCTION
To accelerate MR imaging, many famous deep learning MR image reconstruction methods have been proposed 1,2,3,4 , whose reconstruction time can be reduced to million seconds with encouraging performances achieved. However, most exisiting methods rely on the fully sampled datasets as the groundtruth to do the fully-supervised learning, which may not be accessible for different organs or departments. Hence, weakly supervised learning comes into our view. We want to investigate if weak lables allow us to train a plausible prior network for MR reconstruction. In this work, we proposed a weakly supervised deep prior learning algorithm for multi-coil MR reconstruction, which tries to train a complex UNet with the MR images reconstructed from highly unsampled data with SPIRiT as our coarse-grained label. We have provided some new features into UNet, including introducing complex convolution and replacing upsampling with complex transposed convolutions.THEORY and METHOD
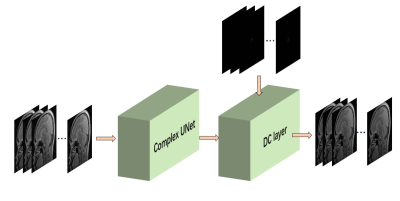
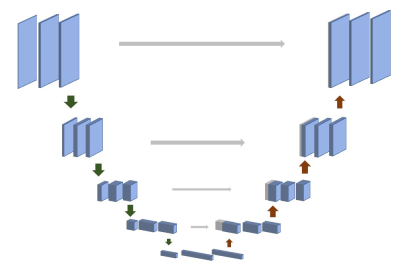
Weakly supervised learning can be mainly classified into three typical types, incomplete supervision , inexact supervision, and inaccurate supervision. In this work, we followed the way of incomplete supervision, it focuses on the situation where exact supervision information are not available, except for imprecise ones. Figure 1 shows the whole process of our proposed algorithm. First of all, SPIRiT was used to reconstruct MR images from undersampled data, which were generated using 1D 4x Uniform mask, as labels of our training process. Then, undersampled images are fed as inputs to train complex UNet. Data consistency layers are also adopted in the network. Figure 2 shows the complete architecture of complex UNet, whose convolution, max pooling layer, up sampling layer were replaced with complex convolution, complex convolution with stride of 2, complex transposed convolution with stride of 2, respectively.RESULTS and DISCUSSION
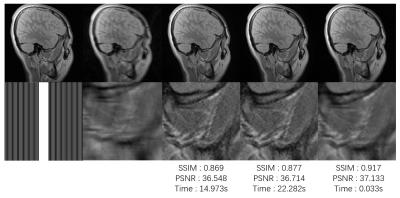
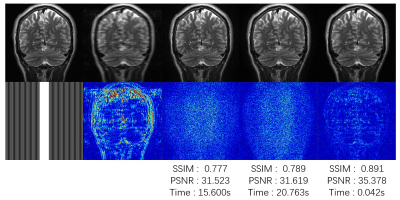
The experimental results are shown in Fig 3 and Fig 4 for the 12-channel T1 and T2 image reconstruction, respectively. The reconstruction time along with the quantitative indicators in PSNR and SSIM, which were calculated between the reconstructed images by different methods and the groundtruth images obtained from the fully sampled k-space data. Fig 3 shows our results controlled the noise in a reasonable range and provide comparable quantitative indicators. But the visual result is a little bit worse compared to the results of GRAPP and SPIRiT. When the image is a little bit noisy, our method can even reconstruct an MR image with higher quantitative indicators compared to the classical reconstruction results. The reason for this phenomenon may be the strong denoising capability of convolutional neural networks.CONCLUSION
In this work, we proposed a weakly supervised deep prior learning algorithm, which trains our desgined complex UNet without accurate labels but still provide encouraging results. Comparing with classical methods, our algorithm can reconstruct image faster and the robustness to noise is slightly stronger 5. And comparing with some other deep learning methods, the needlessness of precise labels is our merit.Acknowledgements
This research was partially supported by the National Natural Science Foundation of China (61601450, 61871371, 81830056), Science and Technology Planning Project of Guangdong Province (2017B020227012, 2018B010109009), Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351), and the Basic Research Program of Shenzhen (JCYJ20180507182400762).References
1. Wang S, Su Z, Ying L, et al. Accelerating magnetic resonance imaging via deep learning[C]//2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). IEEE, 2016: 514-517.
2. Schlemper J, Caballero J, Hajnal J V, et al. A deep cascade of convolutional neural networks for dynamic MR image reconstruction[J]. IEEE transactions on Medical Imaging, 2017, 37(2): 491-503.
3. Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data[J]. Magnetic resonance in medicine, 2018, 79(6): 3055-3071.
4. Zhu B, Liu J Z, Cauley S F, et al. Image reconstruction by domain-transform manifold learning[J]. Nature, 2018, 555(7697): 487.
5. Gong Y, Teng Y, Shan H, et al. Parameter Constrained Transfer Learning for Low Dose PET Image Denoising[J]. arXiv preprint arXiv:1910.06749, 2019.
Figures