3491
Deep Learning based Accelerated MR Image Reconstruction via Transfer Learning1Department of Electrical and Computer Engineering, COMSATS University, Islamabad, Pakistan
Synopsis
In MRI, many deep learning-based solutions often degrade when deployed in different clinical scenarios due to lack of large training datasets. Moreover, the knowledge about the reconstruction problem is constrained to the data seen during training. This paper presents a transfer learning approach to deal with the problems of data scarcity and differences in the source and target domain while reconstructing the MR images using deep learning. Experimental results show successful domain transfer between the source and target datasets in terms of change in magnetic field strength, anatomy and Acceleration Factor (AF).
Introduction
For an ideal performance of neural network, the dataset for training and testing should be drawn from the same domain which may be difficult in MRI due to the differences in scanner hardware, protocols/sequences, under-sampling patterns, training and testing anatomy1. Here we use a transfer learning approach to deal with the differences in the training and testing datasets in the deep learning based accelerated MRI reconstruction. We extract the knowledge about the reconstruction problem from the source domain and apply the learned knowledge to three different target domains with the help of fine tuning.Method
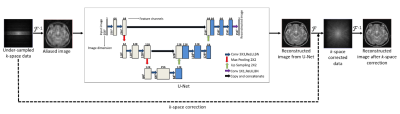
In the first phase, the U-Net is initially trained on a source domain dataset in order to reconstruct the zero filled uniformly under-sampled MR images (AF=4, autocalibration signal (ACS)=12) by the deep learning approach2 shown in Figure 1. For the source domain, 1400 human head Cartesian data (matrix size= 256 X 256) obtained from a 1.5T scanner and a T2-weighted turbo spin-echo pulse sequence3 is used. The training of the U-Net is performed on Python 3.7.1 by Keras using TensorFlow as a backend on Intel(R) core (TM) i7-4790 CPU, clock frequency 3.6GHz, 16 GB RAM and GPU NVIDIA GeForce GTX 780 for approximately 13 hours. RMSprop optimizer is used to minimize the loss function of mean square error.In the second phase of the experiments, the generalization capabilities of the trained U-Net to reconstruct the MR images are investigated for three different target domains: images obtained from 3T scanner4, cardiac images5 and for the images under-sampled by different AFs.
In the third phase of the experiments, the pre-trained U-Net is used to reconstruct the MR images obtained from three different target domains via end-to-end fine tuning6. In end-to-end fine tuning, the knowledge about image reconstruction gained during initial training is transferred to a particular target domain. In this phase, RMSprop optimizer with identical parameters of the initial training is used apart from a lower learning rate.
Results
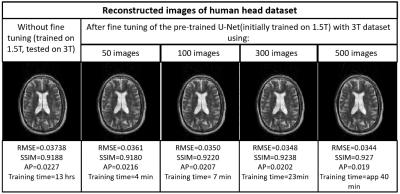
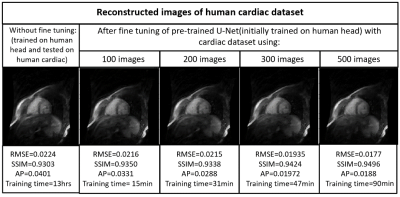
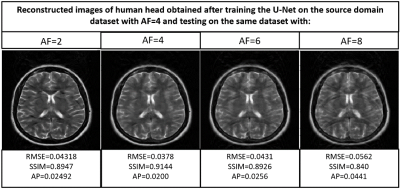
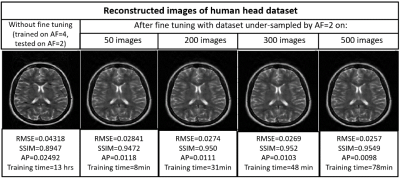
Figures 2,3,4 and 5 show the generalization capabilities of the pre-trained U-Net in terms of the change in magnetic field strength, anatomy and AF. The reconstruction results obtained after fine tuning of the pre-trained U-net on 3T dataset, cardiac dataset and images under-sampled by AF=2 are also shown in Figures 2,3,4 and 5 respectively.Discussion and Conclusions
A neural network initially trained on the human head images obtained from 1.5T scanner can be used to reconstruct brain images obtained from 3T, cardiac images and images under-sampled by different AFs respectively via end-to-end fine tuning with less training time. Hence fine tuning can help us to avoid the training of the neural network every time from the scratch for a particular domain.Acknowledgements
No acknowledgement found.References
1. Daniel Reuckert, 2018, 27th Joint Annual Meeting ISMRM-ESMRMB, lecture notes, Deep learning: Applications in Image Processing, Analysis and Interpretation, Paris Expo Porte de Versailles,Paris,France.
2. Chang Min Hyun et al. Deep learning for under-sampled MRI reconstruction, Phys. Med. Biol. 63 (2018) 135007 (15pp).
3. Loizou et al. (2011), Multiscale amplitude-modulation frequency-modulation (AM–FM) texture analysis of multiple sclerosis in brain MRI images, IEEE Transactions on Information Technology in Biomedicine 15.1 (2010): 119-129.
4. Pamela J. LaMontagne et al, OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and alzheimer’s disease, https://doi.org/10.1016/j.jalz.2018.06.1439.
5. Alexander et al, Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI, volume 12, Issue 3, June 2008, Pages 335-357.
6. Dar, Salman Ul Hassan, et al. "A transfer-learning approach for accelerated MRI using deep neural networks." arXiv preprint arXiv:1710.02615 (2017).
Figures