3489
Predicting the efficacy of chemo-radiotherapy for advanced rectal cancer using support vector machine model and a paired-difference up-sampling strategy under small sample size
Jie Kuang1, Xu Yan2, Gaofeng Shi1, QingLei Shi3, and LI Yang1
1The Fourth Hospital of Hebei Medical University, Shijiazhuang, China, 2Siemens Healthcare, MR Scientific Marketing, Shanghai, China, 3Siemens Healthcare, MR Scientific Marketing, Beijing, China
1The Fourth Hospital of Hebei Medical University, Shijiazhuang, China, 2Siemens Healthcare, MR Scientific Marketing, Shanghai, China, 3Siemens Healthcare, MR Scientific Marketing, Beijing, China
Synopsis
In this study, we proposed a paired-difference analysis (PDA) method to up-sample the training data, which can improve training efficiency of support vector machine (SVM) model with a small sample size. Through optimizing in normalization, dimensional reduction, and features selection steps, a high accuracy was achieved in predicting the efficacy of chemo-radiotherapy for advanced rectal cancer, which means that valuable information may be provided and showed in this clinical situation.
Purpose
With a paired-difference analysis (PDA) method in up-sampling under small sample size, we intended to establish and optimize a support vector machine (SVM) and to evaluate the value of it in predicting the treatment effect of non-metastatic locally advanced rectal cancer (LARC) treated with neoadjuvant chemotherapy-radiation therapy based on radiomics signatures coming from apparent diffusion coefficient (ADC) maps.Materials and Methods
This retrospective study included 55 patients (male 32; female11; age range: 28 to 77 years; mean age: 56.77±12.66) with non-metastatic LARC (adenocarcinoma 38, including 6 cases of poorly differentiation, 30 cases of moderately differentiation, 2 cases of highly differentiation, 4 cases of adenocarcinoma with a small amount of mucinous adenocarcinoma, and 1 case of mucinous adenocarcinoma; pathological stage: low grade 30 cases, high grade 13 cases) scanned from March 2017 to May 2018. All patients were received concurrent chemo-radiotherapy and surgical treatment, with an interval range of 49 to 54 days (mean: 51 days), and all were performed MR examinations at a 3T scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) underwent before and after chemo-radiotherapy treatment within one month. According to curative effect, patients were divided into treatment effective group (TRG0 6 cases; TRG1 8 cases; TRG2 19 cases) and treatment ineffective group (TRG3 10 cases). The inclusion criteria of the study cohort were as follows: (a) MRI scan was performed within 1 week before CRT and within 1-2 weeks after CRT, and the scanned sequence included high-resolution T2WI and DWI (b-values 50 and 800 s/mm2); (b) postoperative pathological data and tumor regression level (TRG) record were complete.Radiomics signatures were extracted using an open source tool named Pyradiomics (https://pyradiomics.readthedocs.io/en/latest/index.html). The PDA method was applied to increase sample size for model training and testing. We gained 378 paired-case differences as the training data set (153/111= positive/negative) and 111 paired-case differences as the independent testing data set (66/48= positive/negative). After comparing the diagnostic performance of models trained with different methods in normalization, dimensional reduction, and features selection, a normal-0-center unit method, a pearson correlation coefficients (PCC), and an recursive feature elimination (RFE) were chosen in these training steps. In this study, a support vector machine (SVM) was used as the classifier, which is an effective and robust classifier to build the model. The kernel function has the ability to map the features into a higher dimension to search the hyper-plane for separating the cases with different labels. Here we used the linear kernel function because it was easier to explain the coefficients of the features for the final model. To prove the performance of the model further, we applied cross validation with 5-folder on the data set.
The performance of the model was evaluated using receiver operating characteristic (ROC) curve analysis. The area under the ROC curve (AUC) was calculated for quantification. The accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were also calculated. All above processes were implemented with FeAture Explorer (FAE, v0.2.5, https://github.com/salan668/FAE) on Python (3.6.8, https://www.python.org/).
Result
We found that the model based on 8 features can get the highest AUC on the validation and test data set. The AUC and the accuracy could achieve 0.819 and 0.891, respectively. In this point, the AUC and the accuracy of the model achieve 0.934 and 0.984 on testing data set, respectively. The sensitivity and specificity were 0.800 and 1.000 on the testing data, with 0.9831 and 1.000 for the NPV and PPV, respectively. The selected features were shown in Table 1, and the ROC curve was shown in Figure 1.Discussions
In order to guarantee the accuracy of the experimental results, we set up strict inclusion criteria. In clinical situations, considering the difficulties of case collection, we employed a paired-difference analysis (PDA) method in up-sampling cases. Using this method, high performance prediction model was achieved using only 55 patient data, which is comparable to that using 378 patient data. Meanwhile, through the subtraction with typical patient data, the variations of radiomics signature may potentially be avoided, which can also improve the accuracy of the model.According to literature’s report and characteristics of different classifiers, a support vector machine (SVM), which is an effective and robust classifier for small samples, was used to build the model. Through comparing the effects to AUC values, a normal-0-center unit method, a PCC method, and a RFE were used in data preparation.
Conclusions
With a small sample size, adopting PDA strategy with the SVM model seems can provide valuable information and showed potential in prediction treatment effect for locally LARC treated with neoadjuvant chemotherapy-radiation therapy.Acknowledgements
References
Figures
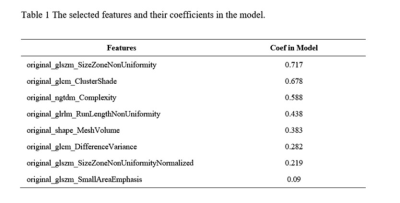
Table 1 The selected features and their coefficients
in the model.
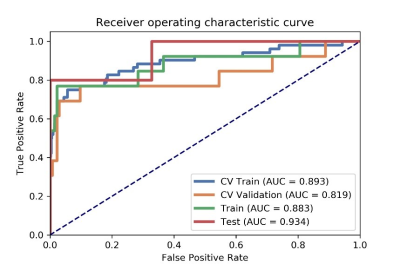
Figure
1 The AUC values of ROC on CV training, CV validation, training and testing
data. CV train: the average of k-1 folder of training data set in k-folder
cross validation; CV validation: the average of k-1 folder of validation data
set in k-folder cross validation.
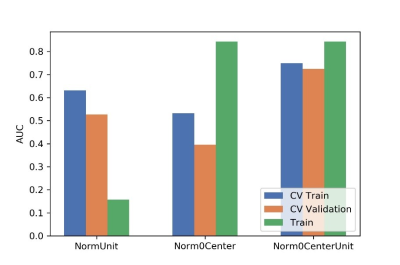
Figure
2 Effects of three normalization method on CV training and CV validation data,
and the corresponding AUCs. CV train: the average of k-1 folder of training
data set in k-folder cross validation; CV validation: the average of k-1 folder
of validation data set in k-folder cross validation.
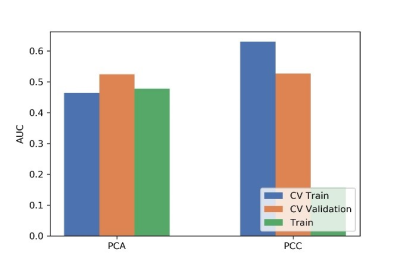
Figure
3 Effects of two dimensional methods on CV training and CV validation data, and
the corresponding AUCs. CV train: the average of k-1 folder of training data
set in k-folder cross validation; CV validation: the average of k-1 folder of
validation data set in k-folder cross validation.
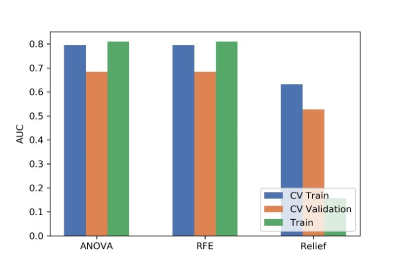
Figure
4 Effects of three feature selective methods on CV training and CV validation
data, and the corresponding AUCs. CV train: the average of k-1 folder of
training data set in k-folder cross validation; CV validation: the average of
k-1 folder of validation data set in k-folder cross validation.
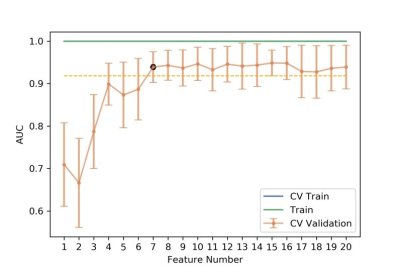
Figure
5 The number of selected features and the corresponding AUCs on CV training and
CV validation, and all training data. CV train: the average of k-1 folder of
training data set in k-folder cross validation; CV validation: the average of
k-1 folder of validation data set in k-folder cross validation.
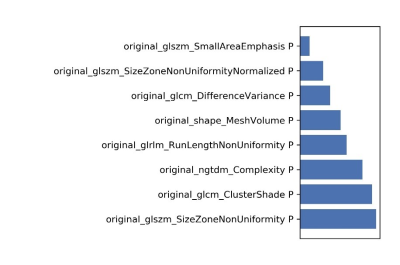
Figure
6 The selected features and their contributions on the model.