3481
Deep Cerebellar Nuclei Segmentation via Semi-Supervised Deep Context-Aware Learning from 7T Diffusion Image1Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States
Synopsis
In this study, we proposed the first deep learning and 7T MR imaging based dentate and interposed nuclei segmentation framework. We introduce dilated dense blocks to effectively encode contextual information on different receptive fields in an encoder-decoder network. Training of the proposed network is optimized with a multi-class hybrid segmentation loss, handling a class imbalance problem. Moreover, a self-training strategy facilitates the training of the proposed network by exploiting auxiliary labels. The proposed framework significantly outperforms an atlas-based deep cerebellar nuclei segmentation tool and state-of-the-art deep neural networks in terms of accuracy and consistency.
Introduction
The cerebellum is primarily not only associated with complex motor, cognitive and linguistic tasks1 but also emotional and perceptual processing.2,3 Of the cerebellum system, deep cerebellar nuclei (DCN) play a pivotal role to form a feedback loop of cerebellar cortex and cerebral cortex.3 Three-dimensional imaging of the DCN is thus a pre-requisite for functional and anatomical studies of the cerebellum and neuro-modulation planning on the relevant region. Moreover, automatic segmentation facilitates subsequent analysis in terms of consistency and efficiency. The existing cerebellum analysis tool4 normalizes the cerebellum anatomy of a specific subject onto a probabilistic atlas (including the DCN) defined on a cerebellum template (SUIT).5 The cerebellum parcellation in the subject space is then performed by inversely applying an estimated warp deformation field to the SUIT atlas. However, such an atlas-based segmentation oftentimes requires additional refinement steps and moreover, atlases do not adequately take into account anatomical variability across the population. Recent advances in 7Tesla (T) MR imaging6–8 and tremendous potential of deep neural networks9–13 enable automatic, fast, and accurate segmentation. In this study, we propose a semi-supervised context-aware deep learning framework against medical data challenges – imbalanced class distribution and limited labeled data - to simultaneously segment deep cerebellar dentate and interposed nuclei using unique 7T diffusion imaging.Methods
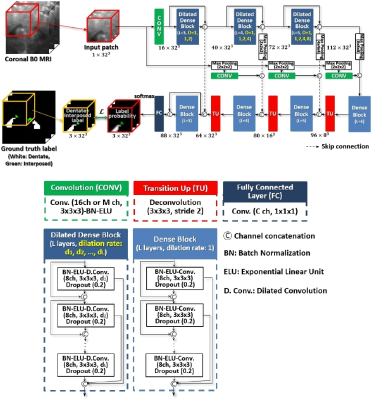
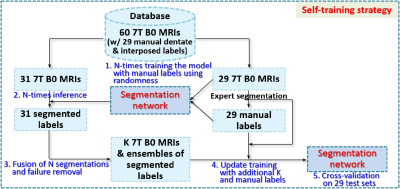
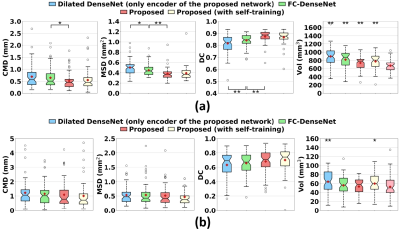
We introduce dilated dense blocks where each layer has a dilated convolution14 to effectively encode contextual information at different scales without consecutive max-pooling and additional complexity in the encoding path of FC-DenseNet12 (Fig. 1). The new encoding path is then integrated into a decoder by adding max-pooling operations in the skip-connections for building a deeper network without a memory burden for input patches. Moreover, we incorporate multi-scale input patches into max-pooled feature maps in the skip-connection to facilitate the learning of local features.15 Further, we propose a self-training strategy that utilizes extra labels from unlabeled data for improving training. As shown in Fig. 2, we train the proposed model N-times with random initialization and manual labels. N labels are predicted on each unlabeled data with N trained models and then fused. This ensemble ensures that the quality of predicted labels is acceptable. We finally re-train the proposed model using a union set of predicted labels and manual labels. The proposed multi-class hybrid segmentation loss ($$$\mathcal{L}$$$) combines Tversky loss16 and focal loss17 and is used during the training, handling the class imbalance problem. The parameters are optimized with Adam (0.001 learning rate). The size of mini-batches is 8 and the number of epochs is 50. The 7T diffusion-weighted MRIs (B0) of 60 subjects were used in this study. The voxel size of the B0 image is 1.25×1.25×1.25mm3. We randomly chose 29 B0 images for validation. Dentate and interposed nuclei were manually labeled on each image and served as ground truth.18 The region of interest on the image was set by linearly co-registering a dentate and interposed nuclei atlas mask of a training image onto a test image. Unlabeled 31 B0 images were used to create extra labels for self-training. We compared the proposed network with SUIT4, popularly used deep neural networks - U-Net10 and FC-DenseNet12. Dice Coefficient (DC)19, center of mass distance (CMD), mean surface distance (MSD) between ground truth and segmented results of each method, and volumes were computed for quantitative analysis.20 For statistical analysis of each measure, a paired t-test was performed on single comparisons. A one-way analysis of variance and Tukey’s honest significance post-hoc test were conducted for multiple comparisons. Five-fold cross-validation on 29 test sets was used for evaluation. 20% of the training data was used as a validation set.Results and Discussion
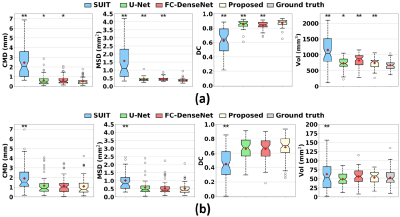
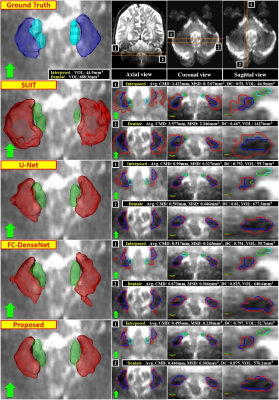
As displayed in Fig. 3, deep learning-based methods significantly outperformed SUIT in every metric for dentate and interposed nuclei segmentation (p<0.001). A large error and variance in SUIT based segmentation might be attributed to uncertainty in registration processes.20 The proposed network produced significantly better dentate segmentation results than other networks (p<0.05). In interposed segmentation, the proposed network also showed better performance than other networks in terms of average errors, which was mostly not statistically significant (p>0.05). The segmentation results of the proposed network were visually closer to the ground truth than other methods (Fig. 4). An ablation experiment proves the effectiveness of the decoder and dilated dense blocks within the proposed network (Fig. 5). The self-training strategy was more effective in interposed nuclei segmentation than dentate segmentation.Conclusion
Volumetric segmentation of deep cerebellar nuclei is a crucial step for functional and neuro-anatomical studies of the cerebellum. In this study, we proposed a novel dentate and interposed nuclei segmentation framework by leveraging a deep neural network and 7T B0 MRI datasets. The proposed network effectively encodes contextual information on different receptive fields using dilated dense blocks. A multi-class hybrid segmentation loss handles a class imbalance problem. Moreover, self-training facilitates the training of the proposed network by distilling data. Experimental results demonstrate that the proposed framework may provide researchers with reliable means for segmenting deep cerebellar nuclei in automatic and efficient ways.Acknowledgements
This work was supported in part by R01-NS081118, R01-NS113746, P50-NS098573, P30-NS076408 and P41-EB027061.References
1. Manto, M. The Cerebellum, Cerebellar Disorders, and Cerebellar Research-Two Centuries of Discoveries. Cerebellum 7, 505–516 (2008).
2. Dennis J. L. G. Schutter & Jack Van Honk. The cerebellum on the rise in human emotion. Cerebellum 4, 290–294 (2005).
3. Baumann, O. et al. Consensus Paper: The Role of the Cerebellum in Perceptual Processes. Cerebellum 14, 197–220 (2015).
4. Diedrichsen, J., Balsters, J. H., Flavell, J., Cussans, E. & Ramnani, N. A probabilistic MR atlas of the human cerebellum. Neuroimage 46, 39–46 (2009).
5. Diedrichsen, J. A spatially unbiased atlas template of the human cerebellum. Neuroimage 33, 127–138 (2006).
6. Abosch, A., Yacoub, E., Ugurbil, K. & Harel, N. An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery 67, 1745–1756 (2010). 7. Cho, Z.-H. et al. Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J Neurosurg 113, 639–647 (2011).
8. Kerl, H. U. et al. The subthalamic nucleus at 7.0 Tesla: Evaluation of sequence and orientation for deep-brain stimulation. Acta Neurochir. (Wien). 154, 2051–2062 (2012).
9. Dolz, J., Desrosiers, C. & Ben Ayed, I. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. Neuroimage 170, 456–470 (2018).
10. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Proc. MICCAI 234–241 (2015).
11. Kamnitsas, K. et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017).
12. Jegou, S., Drozdzal, M., Vazquez, D., Romero, A. & Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. in Proc. CVPR Workshop 11–19 (2017).
13. LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
14. Yu, F. & Koltun, V. Multi-scale context aggregation by dilated convolutions. in Proc. ICLR 1–13 (2016).
15. Lin, T.-Y. et al. Feature Pyramid Networks for Object Detection. in Proc. CVPR 936–944 (2017).
16. Sadegh, S., Salehi, M., Erdogmus, D. & Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. in Proc. MICCAI Workshop (MLMI) 379–387 (2017).
17. He, K., Goyal, P., Girshick, R., Dollar, P. & Lin, T.-Y. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. to be Publ. (2018).
18. Duchin, Y. et al. Patient-specific Anatomical Model for Deep Brain Stimulation based on 7 Tesla MRI. PLoS One 13, 1–23 (2018).
19. Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
20. Kim, J. et al. Automatic localization of the subthalamic nucleus on patient-specific clinical MRI by incorporating 7 T MRI and machine learning: Application in deep brain stimulation. Hum. Brain Mapp. 40, 679–698 (2019).
Figures