3471
High Resolution Sodium MRI Through Manifold Approximation at 3T1Radiology, New York University, New York, NY, United States, 2Katholieke Universiteit Leuven, Leuven, Belgium
Synopsis
We demonstrate the use of manifold approximation for implementing anatomically guided reconstructions for the removal of T2-related blurring from sodium images acquired at 3T.
Introduction
The sodium ion yields the second largest NMR signal of all NMR active nuclei found in the human body. Because of its pivotal role in human physiology, imaging the distribution of the sodium ion during healthy and pathological states is highly desirable. The NMR properties of the sodium nucleus present a number of technical challenges when an accurate assessment of its tissue concentration is desired. Sodium’s very short transverse relaxation rate, in particular, requires the use of techniques capable of ultra-short echo times (1) and leads to image blurring that limits the effective spatial resolution that could be achieved, even when sufficient k-space data have been acquired. Recently, it has been shown that anatomically-guided reconstructions (AGR) using high resolution anatomical information from T1 scans can be used effectively to improve the resolution of low resolution images from multi-modal exams using iterative reconstruction techniques (2). These techniques have also been shown to be amenable for manifold approximation (3) implementations (4) to improve their computational performance thereby providing ease of use. In this paper, we demonstrate the use of such techniques for removing T2-related partial volume effects from sodium images and providing a level of anatomical detail that could not be achieved using existing data acquisition techniques alone.Methods
Data sets were acquired from the brain of normal human volunteers at 3T using a dual-tuned (1H/23Na), eight-channel RF coil and a spectrally-weighted (SW) twisted projection imaging (TPI) readout (6). Acquisition parameters were TE/TR= 0.3/100ms (5-10 Minutes data acquisition time depending on the number of averages used). A high-resolution T1 scan (MPRAGE) was also acquired with a spatial resolution of 1mm isotropic (TE/TR=3/2300ms, TI=900ms). The reconstructed images were used as input to a convolutional neural network (CNN) that had been previously trained to remove partial volume effects on multi-modality (MR/PET) scans with an effective input point-spread-function (PSF) of 4.5mm (4), which is very close to the effective spatial resolution of the sodium scans as a result of the spatial blurring introduced by the T2 decay (see below).Results
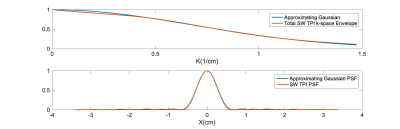
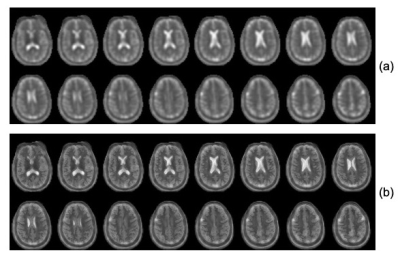
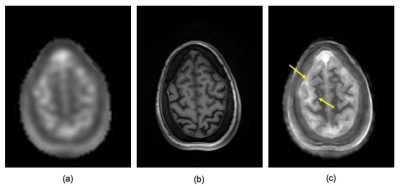
Figure 1 (top) presents the calculated k-space decay envelope for the SW TPI readout used for data acquisition. The envelope was calculated by analytical computation of the time that required to reach each k-space point along the readout and applying the corresponding transverse relaxation decay using experimentally measured rates (T2f=0.008ms, T2s=0.030ms). A Gaussian approximation to this envelope is also shown for easy estimation of the expected PSF (bottom), which agrees well with that obtained through direct Fourier transformation of the calculated k-space decay envelope. Both calculations confirm that the PSF is within the range (4.5-5.5mm) expected to benefit from the AGR approach demonstrated in (2) and (4). Figure 2 presents montages from the original and AGR sodium images from the brain of a human normal volunteer. The images clearly illustrate significant improvements in spatial resolution, which allow differentiating the differences in tissue sodium concentration between gray and white matter that are normally very difficult to assess due to the excessive blurring of the images from the T2 decay. A closer inspection of such improvements is provided in figure 3 where a partition on the superior aspect of the brain is presented. In these images the differences between cerebrospinal fluid (CSF), gray matter and white matter tissue sodium concentrations can be easily assessed and confirm the known differences previously observed on histological specimens (5) despite the fact that the gray matter thickness is very close to the PSF of the input sodium images used by the CNN.Discussion
Sodium images acquired at clinically available magnetic field strengths have long been hampered by severe spatial blurring due to T2 decay. This blurring could be partially ameliorated through a judicious choice of sample density in k-space (6). The resulting spatial resolution is, however, still below the nominal spatial resolution associated with the volume of k-space data being sampled. The use of AGR in this setting provides an unique opportunity to harness the information contained in these inherently quantitative images by reducing the effects of spatial blurring and its associated partial volume effects. The anatomical detail illustrated in figure 3 has not been previously observed in sodium images acquired at 3 Tesla because of the severe blurring experienced during acquisition. Specifically, there is more than a 30% difference in the measured tissue sodium concentration between gray and white matter on these images that, though previously known histologically (5), has been difficult to observe in MR images.Conclusions
Anatomically guided reconstructions can provide a practical means to overcome the deleterious effects of fast transverse relaxation rate on the spatial resolution of sodium images. These gains in spatial resolution have been previously difficult to achieve using existing data acquisition techniques due to the limitations imposed by current gradient technology. These advances are poised to improve our ability to assess the role of sodium MRI in the diagnosis and monitoring of disease in humans.Acknowledgements
This work was supported in part by PHS grants P41 EB017183 and R01R01 NS113517.References
1. Boada F. E., Gillen J. S., Shen G. X., Chang S. Y. and Thulborn K. R.. Fast Three Dimensional Sodium Imaging. Magnetic Resonance in Medicine. 37:706-715, 1997.
2. Schramm G, Holler M, Rezaei A, Vunckx K, Knoll F, Bredies K, Boada F, Nuyts J. Evaluation of Parallel Level Sets and Bowsher's Method as Segmentation-Free Anatomical Priors for Time-of-Flight PET Reconstruction. IEEE Trans Med Imaging. 2:590-603, 2018.
3. Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators. Neural Netw. 2:359–66, 1989
4. Rigie D., Schramm G., Vahle T., Shepherd T., Nuyts J. and Boada F. E. Approximating MRI-Based Anatomically Guided PET Reconstruction with a Convolutional Neural Network. et al., 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 2018, pp. 1-3.
5. Yates A. et al., Postmortem Changes in the Chemistry and Histology of Normal and Edematous Brains. Amer. J. of Pathol, 79:555-564, 1975.
6. Boada F. E., Shen G. X., Chang S. Y. and Thulborn K. R.. Spectrally Weighted Twisted Projection Imaging: reducing T2 signal attenuation effects in fast three dimensional sodium imaging. Magnetic Resonance in Medicine. 38: 1022-1028, 1997.
Figures