3454
Accelerated 4D MRI using joint higher degree total variation and local low-rank constraints (HDTV-LLR)1Harbin Institute of Technology, Harbin, China, 2The First Affiliated Hospital of Harbin Medical University, Harbin, China
Synopsis
Four-dimensional (4D) MRI can provide 3D tissue properties and the temporal profiles at the same time. However, further applications of 4D MRI is limited by the long acquisition time and motion artifacts. We propose a new 4D MRI reconstruction scheme, named HDTV-LLR, by integrating the three-dimensional higher degree total variation and the local low-rank penalties. We demonstrate the benefits of the proposed method using 4D cardiac MRI dataset with undersampling factors of 12 and 16. The proposed method is compared with iGRASP, and schemes using either low-rank or sparsity constraint alone. Results show improved image quality and reduced artifacts.
Introduction
Recently, four-dimensional (4D) MRI has become an emerging developmental technique in radiotherapy treatment planning. However, long scanning time and motion artifacts remain the major barriers preventing the further implementation of 4D-MRI in clinic. With the introduction of compressed sensing, many reconstruction approaches have been proposed for accelerated MRI by exploiting the sparsity in the transform domain. Total variation (TV) regularization scheme has been widely applied in high-dimensional MR data reconstruction1,2. However, TV often leads to patchy artifacts in the recovered images. We have proposed a generalized higher degree total variation (HDTV) scheme3 to reconstruct images from the noisy and undersampled measurements by taking into consideration of all rotations of the directional derivative operators, which overcomes the patchy artifacts caused by TV prior. In recent years, studies have shown that dynamic MRI reconstructions can be significantly improved by enforcing the low-rank constraint4. However, due to the high dimensionality of 4D MR data, the direct application of low-rank regularization is challenging. In order to address the problem, method that promotes local low-rank structure has been proposed5. However, the above mentioned methods are not able to fully exploit the spatio-temporal correlations of the 4D MRI data, thus have limited performance in accelerating 4D MRI. Here, we propose a new algorithm, which incorporates the three-dimensional 2nd degree HDTV component and the local low-rank component into a unified framework. We demonstrate that the proposed method is robust to motion and is able to reconstruct 4D MRI at high undersampling factors.Methods
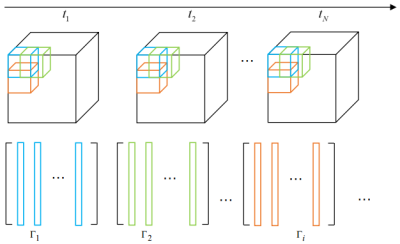
We rewrite the 4D MRI data in a Casorati matrix form $$$\mathbf X \in \mathbb C^{m\times T}$$$, where each column corresponds to the $$$m$$$ voxels in a time frame, and each row corresponds to the time profile of a voxel along $$$T$$$ time frames. The acquisition of the noisy and undersampled MRI measurements $$$\mathbf b$$$ can be modeled as $$$\mathbf b={\cal A}(\mathbf X)+\mathbf n$$$. Here, $$${\cal A}$$$ is the Fourier domain undersampling operator, $$$\mathbf n$$$ is the white Gaussian noise. Due to the high spatio-temporal correlation of the 4D data, the matrix $$$\mathbf X$$$ has the property of low-rank. However, due to the high dimensionality of the 4D MR data, direct application of the low-rank approach is challenging due to the anisotropy of the Casorati matrix, which could also lead to temporal blurring5. Thus, we propose to exploit the local low-rank property by decomposing the data into overlapping blocks in the spatial domain. Each block can thus be rearranged in a low-rank matrix. Fig.1 illustrates the formulation of the matrices based on the local low-rank structure. Moreover, higher degree total variation is demonstrated to considerably improve the image reconstruction performance over standard TV regularization, which enforces the sparsity of the higher order derivatives of high-dimensional data3. Thus, we propose to combine the three-dimensional 2nd order HDTV and the local low-rank penalties to recover the 4D MRI. The problem of recovering the MRI dataset $$$\mathbf X$$$ can be formulated as a constrained optimization function: $$\mathbf X^* =\min\limits_\mathbf X \|{\cal A}(\mathbf X)-\mathbf b \|^2+\lambda_1 \underbrace{\sum\limits_{t=1}^T\int_{\cal S}|\mathbf s^H(\mathbf u){\cal D}(\mathbf X^t)|d\mathbf u}_{\rm{HDTV}}+\lambda_2\underbrace{\sum\limits_{b=1}^K\|\mathbf P_b\mathbf X\|_*}_{\rm{Local\; low-rank}}$$ The optimization function includes the data consistency term, the HDTV, and the local low-rank terms. Here, $$$\mathbf X^t$$$ represents the data volume in the $$$t$$$-th time frame, i.e., the $$$t$$$-th column of $$$\mathbf X$$$. $$$\mathbf s^H(\mathbf u){\cal D}(\mathbf X^t)$$$ is the 2nd degree directional derivative of $$$\mathbf X^t$$$, $$${\cal D}(\mathbf X^t)$$$ is differential operator which provides the 2nd degree partial derivatives of $$$\mathbf X^t$$$, and $$$\mathbf s(\mathbf u)$$$ is the vector of polynomials in the components of the unit directions $$$\mathbf u \in {\cal S}$$$, where $$${\cal S}=SO(3)$$$ is the 3D rotation group, defined with the unit sphere. $$$\mathbf P_b$$$ is the binary matrix operator extracting the $$$b$$$-th block from the data, with the total number of blocks being $$$K$$$. $$$\|\cdot\|_*$$$ is the nuclear norm. $$$\lambda_1$$$ and $$$\lambda_2$$$ are balancing parameters. We use a variable splitting scheme followed by an alternating minimization approach to decouple the original optimization function into three subproblems, where two subproblems can be solved using shrinkage rules, and one quadratic subproblem can be solved with analytic solution. In addition, we adopt the continuation strategy in order to obtain a faster convergence.Results and discussion
The proposed algorithm was applied on a real dynamic 4D cardiac MR dataset of the size 256×256×15×20, provided by the Department of Diagnostic Imaging of the Hospital for Sick Children in Toronto, Canada6. We used the 3D stack-of-stars golden angle radial k-space undersampling scheme7. The proposed HDTV-LLR scheme was compared with 4D-TV, iGRASP7, and the method using local low-rank penalty alone5. Fig.1 and Fig.2 show the recovery results from 12- and 16-fold undersampled measurements, respectively. We observe that the proposed method is able to recover images with sharper edges and lower errors.Conclusion
We propose a HDTV-LLR method, which exploits the spatio-temporal correlation of the 4D MRI data by incorporating the three-dimensional HDTV and local low-rank constraints, in order to reconstruct 4D images at very high undersampling factors. We investigate the utility of the proposed method in recovering a 4D cardiac MRI data from 12-fold and 16-fold undersampled measurements. Results demonstrate the improved performance of HDTV-LLR over existing approaches.Acknowledgements
No acknowledgement found.References
1. Lingala S G, Hu Y, DiBella E, et al. Accelerated dynamic MRI exploiting sparsity and low-rank structure: kt SLR. IEEE transactions on medical imaging. 2011;30(5):1042-1054.
2. Rank C M, Heußer T, Buzan M T A, et al. 4D respiratory motion‐compensated image reconstruction of free‐breathing radial MR data with very high undersampling. Magnetic resonance in medicine. 2017;77(3):1170-1183.
3. Hu Y, Ongie G, Ramani S, et al. Generalized higher degree total variation (HDTV) regularization. IEEE Transactions on Image Processing. 2014;23(6):2423-2435.
4. Christodoulou A G, Zhao B, Zhang H, et al. Four-dimensional MR cardiovascular imaging: Method and applications, 2011. Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2011:3732-3735.
5. Trzasko J, Manduca A, Borisch E. Local versus global low-rank promotion in dynamic MRI series reconstruction, 2011. Proc. Int. Symp. Magn. Reson. Med. 2011;19:4371.
6. Andreopoulos A, Tsotsos J K. Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI. Medical Image Analysis. 2008;12(3):335-357.
7. Feng L, Axel L, Chandarana H, et al. XD‐GRASP: golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing. Magnetic resonance in medicine. 2016;75(2):775-788.
Figures