3442
High-frequency Transform Guided Denoising Autoencoding Prior for Parallel MR Imaging1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China, Shenzhen, China, 2the Department of Electronic Information Engineering, Nanchang University, Nanchang 330031, China, Nanchang, China
Synopsis
Ill-posed inverse problems embodied in parallel imaging remain an active research topic in several decades, with new approaches constantly emerging. Built on the observation that both dictionary learning and conventional sparse coding extract high-frequency component to model, we derived a novel strategy named HDAEP to explore the prior on high-frequency domain on the basis of denoising autoencoding. After the prior is learned from the trained network, the iteratively Gauss-Newton method is employed to jointly estimating the images and coil sensitivities. Experimental results show that the proposed method can achieve superior performances on parallel MRI reconstruction compared to state-of-the-arts.
Theory
High-frequency component can effectively convey most of the semantic information such as texture details of an image [1]. Fig. 1 shows visual characteristics among different high-frequency profiles. As well known, dictionary learning (DL) and conventional sparse coding (CSC) are two popular sparse representation methods. This phenomenon indicates that utilizing high-frequency information facilitates to image reconstruction and it paves a new direction to exploit prior. Therefore, on the basis of denoising autoencoding (DAE) as prior [2], a novel high-frequency strategy acting as a better approximation to the transformation, named HDAEP, is derived.I.Forward and Backward Transform Operators: After reviewing the whole procedure implemented in DL and CSC, we notify that both of them exploit prior in high-frequency domain after decomposing image $$$u$$$ into low-frequency part $$$u_l$$$ and high-frequency part $$$u_h$$$, and they obtain the final reconstruction result by summing the low-frequency domain back. In fact, image decomposition and summation in DL and CSC can be seen as approximations of forward and inverse transform. Specifically, we introduce two operators $$$W$$$ and $$${W^{ - 1}}$$$ to define the forward and inverse transform, i.e., $$$W$$$ transfers the image from the whole image domain to partly high-frequency domain for prior modeling; and $$${W^{ - 1}}$$$ returns the estimation in high-frequency domain back to image domain. Furthermore, we introduce another operator $$$L$$$ which serves as low-pass filtering operator in DL and CSC. We can define $$$W$$$ and $$${W^{ - 1}}$$$ mathematically, i.e., $$$u_h=u-u_l=(I-L)u=Wu$$$, and accordingly, $$$u=u_h+Lu ≈u_h+Lu_h=(I+L) u_h≈(I-L)^{-1} u_h≡W^{-1} u_h$$$. Note that $$${W^{ - 1}}$$$ is obtained by approximated transformation representation.
II.Proposed HDAEP: We inherit the spirit in DL and CSC that using image decomposition and component summation to approximate the forward and inverse transformation. Specifically, there exists two important steps in implementing HDAEP: Forward operator $$$W$$$ and the associated backward operator $$${W^{ - 1}}$$$. Note that in $$$W$$$, a series of high-frequency components is obtained by copying the high-frequency component three times, it is constructive to integrate them for exploiting more detailed prior information. In the approximated backward operator, the updated high-frequency estimation adds with the latest low-frequency components, followed by mean operation to get the final result. We employ the DnCNN [3] architecture as the DAE network $$${A_{{\sigma _\eta }}}$$$ for exploiting high-frequency prior. Correspondingly, the HDAEP prior is denoted as: $$$R(u)=‖W(u)-{A_{{\sigma _\eta }}} (W(u)+\eta)‖^2$$$ where the artificial Gaussian noise $$$\eta$$$ is added to $$$W(u)$$$ as the input of network.
III.Solver for HDAEP: Iteratively Gauss-Newton method (IRGN) [4] is utilized as the solver for HDAEP, and it is demonstrated to improve the performance of auto-calibrating parallel MR imaging mainly due to a better estimation of the coil sensitivity profiles. Mathematically, parallel MR imaging can be considered as a nonlinear inverse problem where the sampling operator $$$F_R$$$ (defined by the k-space trajectory) and the correspondingly acquired k-space data $$$g=(g_1,…,g_N )^T$$$ from $$$N$$$ receiver coils are given, and it is vital to find the spin density $$$u$$$ and the unknown set of coil sensitivities $$$c=(c_1,…,c_N )^T$$$ such that $$$F(u,c) := (F_R (u\cdot c_1 ),…,F_R (u\cdot c_N ))^T=g$$$ holds. Therefore, incorporated with the HDAEP prior, this minimization problem can be solved using IRGN method, i.e., computing in each step $$$m$$$ for given $$$x^m := (u^m,c^m )$$$ the solution $$$dx:=(du,dc)$$$ of the minimization problem is given by $$\min_{dx} \parallel F^{\prime} \left(x^{m}\right)dx+F \left(x^{m}\right)-g\parallel^{2}+\mu_{m}H\left(c^{m}+dc\right)+\lambda_{m}R\left(u^{m}+du\right)$$ where $$$μ_m,λ_m>0,$$$ and setting $$$x^{m+1} := x^m+dx$$$, and $$$λ_{m+1} :=q_λ λ_m$$$ with $$$0<q_{μ,} q_λ<1$$$. Here, $$$F^{\prime}(x^m)$$$ is the Fréchet derivative of $$$F$$$ evaluated at $$$x^m$$$. The term $$$H(c)=‖Hc‖^2=‖h\cdot Fc‖^2$$$ is a penalty on the high Fourier coefficients of the sensitivities and $$$R(u)$$$ regularizes the image part. Specifically, we can tackle the latter one by proximal gradient descent method as illustrated in Fig. 2.
Materials and Methods
All the training and reconstruction experiments were conducted on the PC equipped with Intel Core i7-7700 central processing unit and GeForce Titan XP. The training data set was provided by Shenzhen Institutes of Advanced Technology, the Chinese Academy of Science. All data were scanned from a 3T Siemens’s MAGNETOM Trio scanner using the T1-weighted turbo spin echo sequence. In the basic network, 400 12-channel MR images were used as training data and 100 images for validation. For the purpose of reproducible, source code of HDAEP is available at https://github.com/yqx7150/HDAEP.Results
Fig. 3 compares four images restored using LINDBREG [5] in (b), IGRNTV [4] in (c), SAKE with ESPIRiT [6] in (d), and the proposed HDAEP algorithm in (e) under acceleration rate R=6.7. Obviously, the denoising result of HDAEP method provides much visual improvement and exhibits less blurring artifacts. The PSNR value of HDAEP outperforms LINDBREG with gains of 9.93dB, IGRNTV with gains of 6.47dB, and SAKE with improvement over 5.87dB.Acknowledgements
The study was funded by the National Natural Science Foundation of China (No. 61871206, No. 61661031), the Natural Science Foundation of Jiangxi Province (No. 20181BAB202003).
References
- He Z, Zhou J, Liang D, et al. Learning Priors in High-frequency Domain for Inverse Imaging Reconstruction[J]. arXiv preprint arXiv:1910.11148, 2019.
- Bigdeli S A, Zwicker M. Image restoration using autoencoding priors[J]. arXiv preprint arXiv:1703.09964, 2017.
- Zhang K, Zuo W, Chen Y, et al. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising[J]. IEEE Transactions on Image Processing, 2017, 26(7): 3142-3155.
- Knoll F, Clason C, Bredies K, et al. Parallel imaging with nonlinear reconstruction using variational penalties[J]. Magnetic resonance in medicine, 2012, 67(1): 34-41.
- Wang S, Tan S, Gao Y, et al. Learning joint-sparse codes for calibration-free parallel MR imaging[J]. IEEE transactions on medical imaging, 2017, 37(1): 251-261.
- Uecker M, Lai P, Murphy M J, et al. ESPIRiT—an eigenvalue
approach to autocalibrating parallel MRI: where SENSE meets GRAPPA[J]. Magnetic
resonance in medicine, 2014, 71(3): 990-1001.
Figures

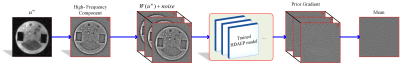
Fig. 2. Schematic flowchart of the proposed HDAEP at iterative reconstruction phase.
