3438
FPGA-based Coprocessor for Real-time GRAPPA Reconstruction (GR-co-RECON)1Electrical and Computer Engineering, Comsats University Islamabad, islamabad, Pakistan, 2Computer Engineering, Khwaja Fareed University of Engineering and Information Technology, Rahim Yar Khan, Pakistan
Synopsis
Data acquisition and reconstruction speed both are crucial for real-time MRI. However, MR image reconstruction speed is highly dependent on the processing capabilities of the hardware platforms (e.g. CPUs, GPUs). Recently, it has been observed that Field Programmable Gate Arrays (FPGA) are a potential candidate to address the computational demands of parallel MRI algorithms. This paper presents the first design effort to implement high performance 32-bit floating-point FPGA-based coprocessor for real-time GRAPPA reconstruction. In-vivo results of 12-channel, 3.0T human-head dataset show that the proposed system speeds up the image reconstruction time up to 106x without compromising image quality.
Introduction
GRAPPA is a well-known pMRI method, which interpolates the under-sampled multichannel k-space data using additionally acquired fully sampled Auto-Calibration Signals (ACS)1. GRAPPA reconstruction process consists of two stages: (i) Calibration (ii) Synthesis. During calibration, the reconstruction coefficients or GRAPPA weight sets are estimated 2 i.e.$$$W=(S_{src}^H S_{src})^{-1} (S_{src}^H S_{tgt} )$$$ 3 ,where $$$S_{src}$$$ and $$$S_{tgt} $$$ are the source and target matrices. In the synthesis phase, estimated weight sets $$$(W)$$$ are used to interpolate the missing data in the acquired k-space i.e. $$$S_{syn}=S_{acq} W$$$ 3, where $$$S_{syn}$$$ represents the missing data in the k-space of each receiver coil and represents the measured data (under-sampled k-space acquired during the accelerated scan).The computational demands of GRAPPA on general purpose computers grow exponentially with an increase in the acquisition parameters e.g. acceleration factor $$$(A_f)$$$, size of convolutional kernel, number of receiver coils $$$(L)$$$ and number of ACS lines 4. This entails a need to offload the processing-intensive tasks of GRAPPA algorithm on FPGAs for fast image reconstruction. In this paper, a 32-bit floating-point FPGA-based coprocessor has been designed for real-time GRAPPA reconstruction. The proposed coprocessor addresses the complexity of GRAPPA reconstruction process by exploiting the existing data-level parallelism in the calibration and synthesis phases.
VIVADO High-Level-Synthesis (HLS) framework has been used to design and implement the proposed coprocessor on Xilinx FPGA development board (ZCU102), which reduces the design effort by raising the abstraction level beyond Register Transfer Logic (RTL) 5. A series of experiments is performed on 12-channel in-vivo human head data to compare the performance of the proposed coprocessor with the CPU-based counterparts.
Methodology
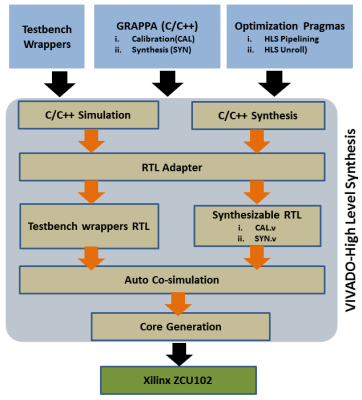
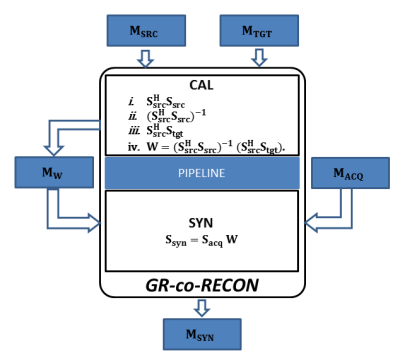
Design flow for the FPGA-based coprocessor for GRAPPA reconstruction (GR-co-RECON) is shown in Figure 1; design specifications of the calibration and synthesis phases are described in two separate modules (CAL and SYN). For high precision 32-bit floating point processing capabilities, float data type is used in the source code. In order to exploit the existing data-level parallelism in the CAL and SYN modules, optimization pragma called “HLS UNROLL” is incorporated in the source code. Moreover, for pipelined data processing between the CAL and SYN modules, optimization pragma called “HLS PIPELINE” is used. The source code of GRAPPA reconstruction algorithm along with optimization pragmas is subjected to VIVADO-HLS framework that performs advanced platform-based code transformations and optimizations to generate an optimized synthesizable RTL. In order to verify the correctness of the synthesized RTL, automatic co-simulation is performed using C++ test bench wrappers. Thereafter, the synthesized RTL for GRAPPA reconstruction is implemented as a coprocessor on Xilinx ZCU102 FPGA development board using VIVADO Design suite.Figure 2 shows a complete flow of operations for GRAPPA reconstruction using the proposed coprocessor (i.e. GR-co-RECON) which starts its operation by reading the source and target matrices form respective memory banks i.e. $$$M_{SRC}$$$ and $$$M_{TGT}$$$ . CAL module solves a large over-determined system of linear equations to estimate the GRAPPA weight sets $$$(W)$$$ and updates the reconstruction coefficients in the memory bank $$$(M_{W})$$$. SYN module reads $$$W$$$ and $$$S_{acq}$$$ from the memory banks (i.e. $$$M_{W}$$$ and $$$M_{ACQ}$$$ respectively) and recovers the missing data in the under-sampled region of the k-space as shown in Figure 2.
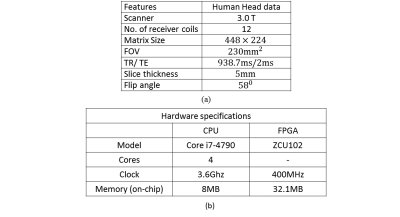
In order to validate the performance of the proposed GR-co-RECON, several experiments are performed on 12-channel in-vivo human head dataset. The data acquisition details and hardware platform specifications are presented in Table 1. For performance comparison between the proposed GR-co-RECON and the CPU-based counterparts, reconstruction time and accuracy (in terms of SNR and RMSE) are evaluated for 32-ACS lines and different acceleration factors (e.g. $$$A_f=2,4$$$).
Results and Discussion
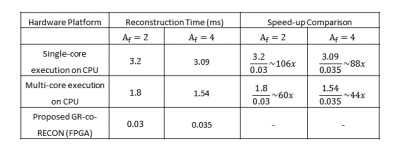
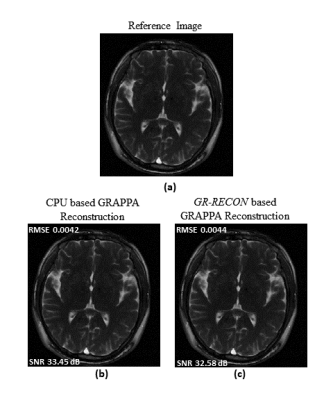
Table 2 shows a performance comparison between the proposed GR-co-RECON and the CPU-based counter parts. For visual comparison, the CPU and GR-co-RECON reconstructed images for 12-channel in-vivo human head data along with SNR and RMSE values are shown in Figure 3. The results show that the proposed GR-co-RECON withstands the reconstruction quality of GRAPPA reconstruction algorithm while demonstrating a maximum speed-up of 106x over the CPU-based counterparts.Conclusion
This paper presents the design and implementation details of FPGA-based coprocessor for GRAPPA reconstruction (GR-co-RECON); optimized for an efficient and high-quality image reconstruction. When compared with the CPU-based counter parts, GR-co-RECON significantly reduces the reconstruction time (speed up factor upto 106x) while maintaining visual quality of the reconstructed image.Acknowledgements
No acknowledgement found.References
1. Griswold, Mark A., et al. "Generalized autocalibrating partially parallel acquisitions (GRAPPA)." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 47.6 (2002): 1202-1210.
2. Breuer, Felix A., et al. "General formulation for quantitative G‐factor calculation in GRAPPA reconstructions." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 62.3 (2009): 739-746.
3. Inam, Omair, et al. "Iterative Schemes to Solve Low-Dimensional Calibration Equations in Parallel MR Image Reconstruction with GRAPPA." BioMed research international 2017 (2017).
4. A. C. S. Brau,P. J.Beatty, S. Skare, and R.Bammer, “Comparison of reconstruction accuracy and efficiency among autocalibrating data-driven parallel imaging methods,”. Magnetic Resonance in Medicine, vol. 59, no. 2, pp. 382–395.
5. Cong, Jason, et al. "High-level synthesis for FPGAs: From prototyping to deployment." IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems 30.4 (2011): 473-491.
Figures