3437
Advanced New Linear Predictive Reconstruction Methods for Simultaneous Multislice Imaging1Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Martinos Center for Biomedical Imaging, Charlestown, MA, United States
Synopsis
Many autocalibrated parallel imaging reconstruction methods are based on linear-predictive/autoregressive principles, including noniterative GRAPPA-type interpolation methods, iterative SPIRiT-type annihilation methods, and structured low-rank matrix methods like PRUNO and Autocalibrated LORAKS. In principle, all of these approaches could be adapted for simultaneous multislice (SMS) reconstruction. However, in practice, GRAPPA-type SMS methods are popular, but there has been limited exploration of more advanced annihilation-based or structured low-rank matrix SMS methods. In this work, we adapt and evaluate these advanced approaches for SMS reconstruction. Results demonstrate that these advanced approaches can offer substantial improvements over simpler GRAPPA-type methods when applied to SMS.
Introduction
Over the past 20 years, autocalibrated linear-predictive (ALP) reconstruction methods have become very prevalent in parallel imaging. Some of the most popular ALP approaches include GRAPPA-type interpolation methods1, SPIRiT-type annihilation methods2, and autocalibrated structured low-rank methods like PRUNO3 and Autocalibrated LORAKS4. While these different approaches all have different formulations and performance characteristics, they are all closely related and can be understood under a unified theoretical framework5.Although these ALP methods can easily be generalized to simultaneous multislice (SMS) reconstruction problems6,most existing SMS ALP methods are of GRAPPA-type1,7-8. Annihilation-based (SPIRiT-type) and structured low-rank SMS ALP methods have only recently begun to be explored9,10, with only limited comparisons against GRAPPA-type SMS methods. In addition, the structured low-rank ALP methods proposed so far for SMS9 do not yet take full advantage of some of the powerful relationships that have been used effectively in ALP parallel imaging3-5. Based on the previous parallel imaging literature, the advanced annihilation-based and structured low-rank approaches would be expected to outperform the GRAPPA-type approaches, and are therefore very promising to investigate.
We undertake such a task in this work, by adapting and evaluating annihilation-based and structured low-rank ALP methods for SMS, with comparisons against GRAPPA-type SMS ALP methods.
Methods
ReconstructionWe implemented and compared two existing SMS ALP methods and two new ones. The existing methods included a GRAPPA-type method (Split Slice-GRAPPA8) and an annihilation-based method (Slice SPIRiT10), while the new methods are simple adaptations of existing ALP structured low-rank methods3,4,11. The first new method, which we call SMS AC-LORAKS, follows the same basic principles as PRUNO3 and Autocalibrated LORAKS4, and solves $$\hat{\mathbf{k}} = \arg\min_{\mathbf{k}} \sum_{\ell=1}^{L}\|\mathcal{P}_C(\mathbf{k}_\ell)\mathbf{N}_\ell\|_F^2 \mathrm{\,\,\,\,\, subject\, to\,\,\,\,\, } \mathcal{A}(\mathbf{k})=\mathbf{d},$$ where $$$L$$$ is the number of simultaneous slices; $$$\mathbf{d}$$$ is the vector of observed multichannel SMS data; $$$\mathbf{k}_\ell$$$ is the multichannel data for the $$$\ell$$$th slice (unknown, to-be-estimated) and $$$\mathbf{k}$$$ is the concatenation of all the $$$\mathbf{k}_\ell$$$ values; $$$\mathcal{A}$$$ models the SMS acquisition procedure (including both SMS encoding and in-plane acceleration); $$$\mathcal{P}_C(\cdot)$$$ is an operator that constructs a convolution-structured matrix from the multichannel k-space data3,4 (i.e., the LORAKS “C-matrix”12); and $$$\mathbf{N}_\ell$$$ is the nullspace matrix for the $$$\ell$$$th slice, which is learned from autocalibration (ACS) data3,4.
The second new method follows the same basic principles described above, except that we are inspired by the modified Autocalibrated LORAKS approach of Ref. 11 to add an additional nonconvex structured low-rank regularization term that automatically identifies and imposes support, phase, and parallel imaging constraints. This new approach, which we call Regularized SMS AC-LORAKS, solves $$\hat{\mathbf{k}} = \arg\min_{\mathbf{k}} \sum_{\ell=1}^{L}\|\mathcal{P}_C(\mathbf{k}_\ell)\mathbf{N}_\ell\|_F^2 + \lambda \sum_{\ell=1}^{L}J(\mathcal{P}_S(\mathbf{k}_\ell)) \mathrm{\,\,\,\,\, subject\, to\,\,\,\,\, } \mathcal{A}(\mathbf{k})=\mathbf{d},$$ where $$$J(\cdot)$$$ is a nonconvex penalty that promotes low rank; and $$$\mathcal{P}_S(\cdot)$$$ is an operator that constructs an alternative convolution-structured matrix that can take advantage of the relationships between k-space samples on opposite sides of k-space (i.e., the LORAKS “S-matrix”12) that can arise when an image has smooth phase characteristics.
Evaluation
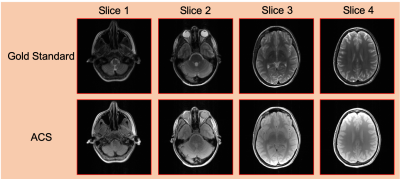
The SMS ALP methods were evaluated using in vivo 16-channel spin-echo brain data13, which was retrospectively slice-combined and retrospectively undersampled. We emulated an SMS experiment with CAIPI14 sampling, multiband factor=4, and uniform in-plane acceleration factors of R=1, 2, and 3. The ACS data was obtained from the same slices with different sequence parameters (yielding different contrast for the ACS data compared to the undersampled datasets being reconstructed). The gold standard and ACS datasets are shown in Fig. 1.
Results
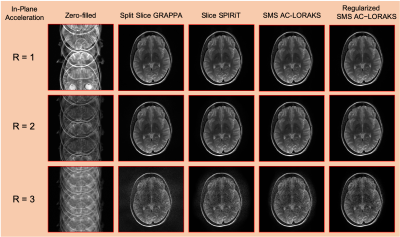
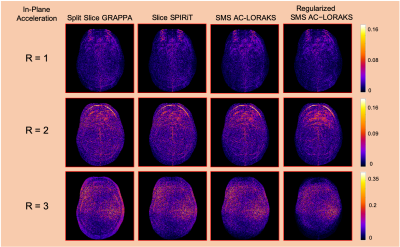
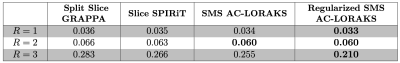
Reconstruction results are shown in Fig. 2 and error images are shown in Fig. 3 for the different cases, with quantitative errors reported in Fig. 4. The results demonstrate that all methods work well for this data when there is no in-plane acceleration, while differences begin to emerge as the acceleration factor increases. We observe that Split Slice GRAPPA has the worst performance among all of these approaches at high-acceleration factors. Slice SPIRiT outperforms Split Slice GRAPPA, but is outperformed by SMS AC-LORAKS, which is itself outperformed by Regularized SMS AC-LORAKS.Discussion and Conclusions
In this work, motivated by the success of advanced ALP methods for conventional (non-SMS) parallel imaging reconstruction, we adapted and evaluated SPIRiT-type and structured low-rank ALP methods for SMS reconstruction. Consistent with the previous parallel imaging literature, our results demonstrate that these kinds of approaches can offer substantial advantages in SMS applications compared to GRAPPA-type methods when the acceleration factor is high. As such, we believe these reconstruction approaches are worth considering as a replacement for previous SMS reconstruction methods in applications that demand high acceleration factors. In addition, all of these methods can easily be combined with additional constraints and/or regularization penalties to improve image reconstruction quality even further.Acknowledgements
This work was supported in part by research grants NSF CCF-1350563, NIH R01 NS074980, NIH R01 NS089212, and NIH R01 MH116173.References
[1] Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. "Generalized autocalibrating partially parallel acquisitions (GRAPPA)." Magnetic Resonance in Medicine 47:1202-1210, 2002.
[2] Lustig M, Pauly JM. "SPIRiT: iterative self‐consistent parallel imaging reconstruction from arbitrary k‐space." Magnetic Resonance in Medicine 64:457-471, 2010.
[3] Zhang J, Liu C, Moseley ME. "Parallel reconstruction using null operations." Magnetic Resonance in Medicine 66:1241-1253, 2011.
[4] Haldar JP. "Autocalibrated LORAKS for fast constrained MRI reconstruction." Proc. IEEE International Symposium on Biomedical Imaging, pp. 910-913, 2015.
[5] Haldar JP, Setsompop K. "Linear Predictability in MRI Reconstruction: Leveraging Shift-Invariant Fourier Structure for Faster and Better Imaging." IEEE Signal Processing Magazine, 2020. (Preprint: arXiv:1903.03141)
[6] Barth M, Breuer F, Koopmans PJ, Norris DG, Poser BA. "Simultaneous multislice (SMS) imaging techniques." Magnetic Resonance in Medicine 75:63-81, 2016.
[7] Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. "Blipped‐controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g‐factor penalty." Magnetic Resonance in Medicine 67:1210-1224, 2012.
[8] Cauley SF, Polimeni JR, Bhat H, Wald LL, Setsompop K. "Interslice leakage artifact reduction technique for simultaneous multislice acquisitions." Magnetic Resonance in Medicine 72:93-102, 2014.
[9] Park S, Park J. "SMS‐HSL: Simultaneous multislice aliasing separation exploiting Hankel subspace learning." Magnetic Resonance in Medicine 78:1392-1404, 2017.
[10] Sun C, Yang Y, Cai X, Salerno M, Meyer CH, Weller D, Epstein FH. "Non‐Cartesian slice‐GRAPPA and slice‐SPIRiT reconstruction methods for multiband spiral cardiac MRI." Magnetic Resonance in Medicine 2019. Early View.
[11] Lobos RA, Kim TH, Hoge WS, Haldar JP. "Navigator-free EPI Ghost Correction with Structured Low-Rank Matrix Models: New Theory and Methods." IEEE Transactions on Medical Imaging 37:2390-2402, 2018.
[12] Haldar JP. "Low-rank modeling of local k-space neighborhoods (LORAKS) for constrained MRI." IEEE Transactions on Medical Imaging 33:668-681, 2013.
[13] Bilgic B, Kim, TH, Liao C, Manhard MK, Wald LL, Haldar JP, Setsompop K. "Improving parallel imaging by jointly reconstructing multi‐contrast data." Magnetic Resonance in Medicine 80:619-632, 2018.
[14] Breuer FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM. "Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi‐slice imaging." Magnetic Resonance in Medicine 53:684-69, 2005.
Figures