3418
3DANCE VDS: Three-points Dixon golden Angle with off-resoNance CorrEction Variable Density Spiral1Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States
Synopsis
3DANCE is a sequence designed to be a good candidate for a whole-body sequence for multiple applicaitons. Phantom and in vivo experiments demonstrated a good spatial resolution and consistent ability to separate water and fat tissues. Off-resonance correction of the water and fat images have showed good results in vitro but needs more testing for in vivo datasets. Future steps include improving the robustness of the methods and testing for motion tolerance.
Introduction
Current Magnetic Resonance Imaging (MRI) scenario supposes being inside the scanner lying still for an average of 45 minutes per body part, subjected to loud noises and sometimes even holding the breath for periods longer than 30 seconds. The MR community has to devote efforts to overcome these issues to ensure an optimum patient experience. A good candidate sequence to counteract some of these issues should possess features such as fast acquisition, certain degree of motion tolerance and body quantification capabilities. 3DANCE VDS conceptually meets these requirements by implementing 1. Dixon water-fat separation1, 2. spiral variable density k-space sampling pattern, 3. motion tolerance by the continuous acquisition of spiral shots at the golden angle2 and the possibility to deblur the images by off-resonance correction without the need of an external field map3. Although the sequence is still in developing mode, we have started testing some of its features. The aim of this work is to provide a status update on the feasibility of the method in terms of water-fat separation, image quality and deblurring.Methods
3-DANCE VDS sequence consists of a steady-state free precession-based sequence with a VDS trajectory, this feature allows for a more efficient k-space coverage and short total acquisition times. Specifically, the acquisition sequence has 54 shots with a center oversampling factor of 9. Dixon technique is achieved by introducing TE delays every three spiral shots. The echo of the first shot is acquired at TE, the second one at TE+dTE and the third one at TE+2dTE. This is repeated sequentially through all the shots. After this, we combine the shots with the same echo time together to form three lower resolution images or echos. These images are used for the calculation of the field map following the method described in ref3. The phase shift between the first and second echos are used for the calculation of the water and fat only images using the formulas described in ref.5 Water and fat images are sampled again along the acquisition trajectory to obtain a deblurred version of them using the previously calculated field map as a guide using frequency-segmented CPR. 4Two types of experiments were carried out: in vitro and in vivo. For the in vitro case, image resolution was assessed using the ACR6 phantom whereas tissue separation was validated and optimized for water and oil phantoms. In vivo experiments were performed on the thighs of a healthy volunteer. Sequence design was performed using Pulseq7,8, an open-source and vendor-agnostic framework that allows for rapid implementation and execution on the hardware. The sequence parameters are FA=30 degrees, TE/TR=4.6/25 ms, dTE=1.22ms, FOV=384 cm, in-plane resolution of 2x2 mm2 and slice thickness of 3 mm. Testing and validation of the sequence were performed on a 3T Siemens Prisma system using the body coil. Image reconstruction utilizes python code for non-uniform Fourier transform.9
Results and discussion
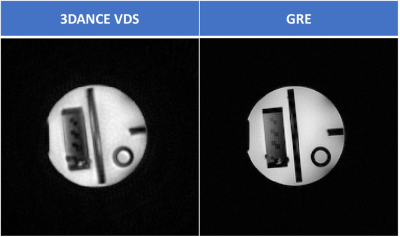
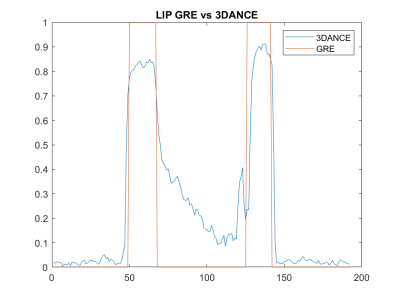
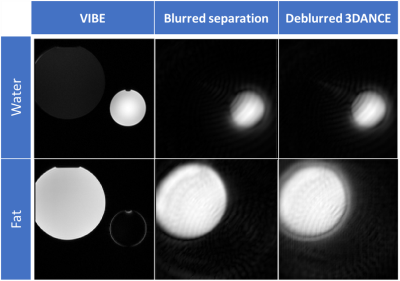
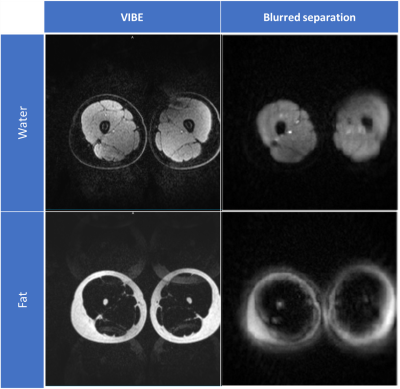
Figure 1 compares the image resolution of 3DANCE with a gradient echo acquisition. The main difference is the off-resonance blurring, however, comparison of line intensity profiles in figure 2 shows a tight match. Only edges are not as sharp. Figure 3 shows the agreement on tissue separation for Voxel Interpolation Breath-Hold Examination (VIBE) and 3DANCE before and after deblurring in vitro. The phantom edges are sharper after off-resonance correction but gradient delay artifact shows as a halo surrounding them. Figure 4 show Dixon performance in vivo. While tissue discrimination is good the resolution is poor due to blurriness. The field of view was large and impacted by the non-linearity of the gradients. These r results should be improved by off-resonance correction. Golden angle is also a feature of this sequence. It will increase the inherent motion tolerance of spirals. Future efforts will be devoted to verify it experimentally. The benefits that this sequence could bring are key for its application on fields such as MR safety, water-fat tissue measurements and health screening of large populations.Conclusion
3DANCE VDS has been demonstrated to perform good water-fat discrimination. The use of spirals result on blurring of the images but it can be corrected using off-resonance correction methods without the need for an external field map. Future steps will be devoted to test how well the golden angle provides motion tolerance and to make field map calculation more robust, allowing for optimal deblurring.Acknowledgements
1. Zuckerman Institute Technical Development Grant for MR, Zuckerman Mind Brain Behavior Institute,Grant Number: CU-ZI-MR-T-0002; PI: Geethanath;
2. Zuckerman Institute Seed Grant for MR studies, Zuckerman Mind Brain Behavior Institute,Grant Number:CU-ZI-MR-S-0007; PI: Geethanath;
References
1. G. H. Glover, “Multipoint dixon technique for water and fat proton and susceptibility imaging,” J. Magn. Reson. Imaging, vol. 1, no. 5, pp. 521–530, 1991.
2. S. Winkelmann, T. Schaeffter, T. Koehler, H. Eggers, and O. Doessel, “An optimal radial profile order based on the golden ratio for time-resolved MRI,” IEEE Trans. Med. Imaging, vol. 26, no. 1, pp. 68–76, 2007.
3. K. S. Nayak, C. M. Tsai, C. H. Meyer, and D. G. Nishimura, “Efficient off-resonance correction for spiral imaging,” Magn. Reson. Med., vol. 45, no. 3, pp. 521–524, 2001.
4. D. C. Noll, J. M. Pauly, C. H. Meyer, D. G. Nishimura, and A. Macovskj, “Deblurring for non‐2D fourier transform magnetic resonance imaging,” Magn. Reson. Med., vol. 25, no. 2, pp. 319–333, 1992.
5. H. Moriguchi, J. S. Lewin, and J. L. Duerk, “Dixon Techniques in Spiral Trajectories With Off-Resonance Correction: A New Approach for Fat Signal Suppression Without Spatial-Spectral RF Pulses,” Magn. Reson. Med., vol. 50, no. 5, pp. 915–924, 2003.
6. Chen, C. C., Wan, Y. L., Wai, Y. Y., and Liu, H. L., (2004), Quality assurance of clinical MRIscanners using ACR MRI phantom: preliminary results. Journal of digital imaging, 17(4): 279-284.
7. Layton, K. J., Kroboth, S., Jia, F., Littin, S., Yu, H., Leupold, J., Nielsen, J., Stöcker, T. and Zaitsev,M. (2017), Pulseq: A rapid and hardware‐independent pulse sequence prototyping framework.Magn. Reson. Med., 77: 1544-1552. doi:10.1002/mrm.26235
8. Ravi et al., (2019). PyPulseq: A Python Package for MRI Pulse Sequence Design. Journal of OpenSource Software, 4(42), 1725, https://doi.org/10.21105/joss.01725
9. Lin, Jyh-Miin (2018), Python Non-Uniform Fast Fourier Transform (PyNUFFT): AnAccelerated Non-Cartesian MRI Package on a Heterogeneous Platform (CPU/GPU). Journal ofImaging, 4(3): 51.
Figures