3383
Clinical Image Quality Evaluation For Field Strength and Contrast Independence of Deep Learning Reconstruction1Canon Medical Systems USA, Inc, Tustin, CA, United States, 2Canon Medical Systems USA, Inc., Tustin, CA, United States
Synopsis
dDLR algorithms with two path CNN architecture that are designed to be noise adaptive are robust against differences in image contrast and field strength. This study shows clinical image quality improvement on 1.5T brain and knee datasets using an algorithm trained on 3T data.
Introduction
Denoising approaches with deep learning based reconstruction (dDLR) have been shown to be adaptive to various noise levels and contrasts of MR images (1,2). Following training and validation at 3T with pairs of high SNR ground truth and noisy images (Ground truth + artificial gaussian noise, which SD is [0 20](%) of max. SI), dDLR has been reported to outperform block matching and 3D filtering (BM3D)(2). In phantom and volunteer testing, dDLR significantly reduced image noise while preserving image quality for brain MR images on 3T(3). dDLR uses a two path DCNN architecture: 1- feature extraction and multiple feature conversion layers and 2 – a separate pathway for the zero-frequency component of the discrete cosine transformation (DCT). Separation of this zero-frequency component from the feature extraction layer maintains the image contrast. Furthermore, a single neural network is trained by the entire training set containing a variety of contrasts , therefore even the contrast difference in the sample images have no impact on the result, since noise is being removed regardless of contrast weighting. Given the various types contrast possibilities of images and levels of noise in MR, as well as changes in tissue characteristics at 1.5T compared to 3T, the purpose of this study was to verify that dDLR can be applied to 1.5T based on its architectural design. The goal of this study was to determine whether the dDLR algorithm trained on 3T data can improve image quality on clinical 1.5T brain and knee images on a qualitative scale.Methods
Twenty clinical brain raw data studies and twenty clinical knee raw data studies were collected with permission from 1.5T MRI scanners (Vantage Orian, Canon Medical Systems, Tochigi, Japan), anonymized, reconstructed using dDLR and 3 standard clinical denoising and gain algorithms (NL2, GA43 and GA53), and randomized. The anonymized and randomized brain and knee clinical datasets were uploaded to an online Dicom viewer to be reviewed blindly by 3 Neuro radiologists and 3 MSK radiologists, respectively. Each of the 20 brain studies included the following sequences: Diffusion, T1, T2, T2*, MP-Rage, FLAIR, TOF, and post Gd. Each of the 20 knee studies included four copies of the following sequences: PD, PD FS, T1, T2 STIR, T2, and T2 FS. Each radiologist viewed and qualitatively evaluated each series based on the appearance of image sharpness, image contrast, SNR, image noise, image noise texture homogeneity, lesion/pathology conspicuity and overall image quality according to a 1-5 Likert scale (4). The Friedman test was used to determine overall statistical significant difference within the group, followed by separate post-hoc Wilcoxon signed-rank tests on the different combinations of groups with a Bonferroni adjustment for a significance level of 0.017 (0.05/3). SPSS software (IBM) was used for statistical analysis.Results
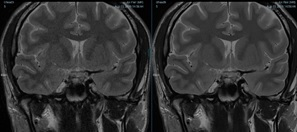
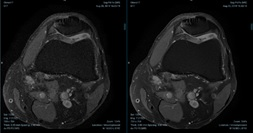
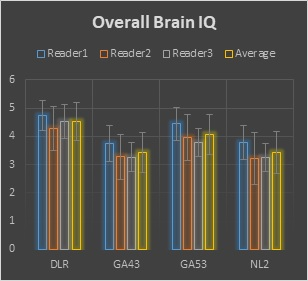
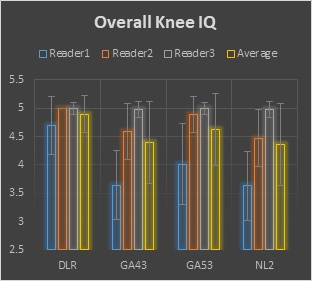
Representative brain and knee images used in the evaluation are shown in Figures 1 and 2. For the 1.5T brain evaluation, dDLR was preferred by the radiologists compared to NL2, GA43, and GA53 according to the higher average score of DLR in all review categories. Significant differences were found for all comparisons (p<0.0001). dDLR scored higher than the other filters in overall image quality, Image sharpness, SNR, contrast, noise, noise texture homogeneity, and lesion conspicuity. Average brain image quality scores for each filter and reviewer is shown in Figure 3. For the knee evaluation, dDLR was preferred by the radiologists compared to NL2 in all review categories according to the higher average score of dDLR (p<0.0001). Between DLR:GA53 and DLR:GA43, strong preferences for DLR was observed in overall image quality, sharpness, SNR, noise, noise texture homogeneity, and lesion conspicuity (p<0.0001). Average brain and knee overall image quality scores for each filter and reviewer is shown in Figures 3 and 4. All images were diagnostic.Discussion
The dDLR training data set comprised of pairs of a high-SNR target and a noisy input images, optimized through an offline training process, has learned to differentiate signal from noise and can then be applied to denoising input images regardless of contrast and field strength. dDLR can handle the differences between various contrast weightings at 3T (3), where the contrast differences are large. This study shows that dDLR can be applied to 1.5T with improved image quality. Any subtle contrast differences in 1.5T images are not affected by the dDLR algorithm. The results of this clinical evaluation indicates that the noise adaptive CNN algorithm and two path architecture make it possible to apply dDLR to clinical 1.5T data with improved image quality.Conclusion
The noise adaptive dDLR algorithm with two path architecture trained on 3T brain and knee data can successfully be applied to 1.5T brain and knee clinical images. Further clinical studies will be conducted to explore further possible clinical benefits of dDLR.Acknowledgements
No acknowledgement found.References
1. Isogawa K et al. Noise level adaptive deep convolutional neural network for image denoising. Proceedings of ISMRM Paris, 2018; 2797.
2. Shinoda K et al. Deep Learning Based Adaptive Noise Reduction in Multi-Contrast MRI; ISMRM 2019 #4701
3 Kidoh M et al. Deep Learning Based Noise Reduction for Brain MR Imaging: Tests on Phantoms and Healthy Volunteers. Magn Reson Med Sci doi:10.2463/mrms.mp.2019-0018
4 Cheng JY et al. (2019) Compressed Sensing: From Research to Clinical Practice with Data-Driven Learning (2019).
Figures