3382
Design of deep neural network in time-phase encoding plane for compressed sensing cardiovascular CINE MRI1Kwangwoon University, Seoul, Republic of Korea
Synopsis
We build a deep neural network in time-phase encoding plane (t-y) for compressed sensing cardiovascular CINE MRI. Previously neural networks were developed in cross-sectional image planes (x-y). A hierarchical convolutional neural network (CNN), known as U-net is used. By adopting the t-y plane, instead of the x-y plane, simultaneous restoration in time and space is effectively achieved. By computer simulation, the proposed deep neural network based on the t-y plane shows better cross-sectional images, clearer temporal profiles, and less normalized mean square errors compared to that of the x-y plane.
Introduction
Deep artificial neural network has been successfully applied to various fields including image segmentation, classification, and reconstruction. U-net, a hierarchical convolutional neural network by varying kernel and channel sizes has widely been used due to its superior performance.1 Although cardiovascular CINE imaging has four-dimensional characters, deep neural network is mostly constructed for two-dimensional input and output due to exponentially increasing complexity with higher dimensions. Previous neural networks were developed in cross-sectional image planes (x-y), where learning about spatial restoration is made, but learning about temporal restoration is difficult to achieve. In this study, neural network is established in time-phase encoding plane (t-y) directly related to compressed sensing. The performance of the neural network in the t-y plane is compared to that of the x-y plane for cardiovascular CINE MRI.Methods
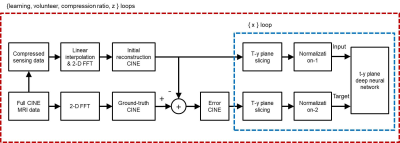
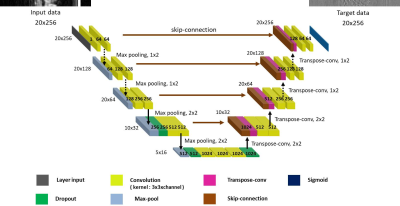
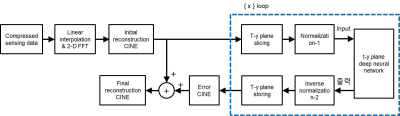
Block diagram for training neural network in the t-y plane is shown in Fig.1. For supervised learning, ‘full CINE MRI data’ is acquired without compression at a 3.0T MRI system (Siemens) with a balanced SSFP sequence for 8 healthy volunteers.2 The imaging parameters are as follows. TR = 3.88ms, TE = 1.94ms, VPS = 8, pixel resolution = 1.37mm x 1.37mm x 8mm, number of slices = 12, and number of frames = 20. Training and test data are prepared exclusively (volunteers 1 - 4 for training data and 5 - 8 for test data). Reconstruction of full CINE data by 2-D FFT is assumed to be ‘ground-truth CINE’ images. ‘Compressed sensing data’ are generated by subsampling of the full data with compression ratios (CR) of 2, 3, and 4. The missing data are first interpolated by the measured data of adjacent frames, which are then 2-D Fourier transformed to make ‘initial reconstruction CINE’ images.3 The ground-truth images are subtracted by the initial reconstruction images to make ‘error CINE’ images. The initial reconstruction and error CINE images are sliced into t-y planes, which are fed into the ‘t-y plane deep neural network’ as the input and the target after normalization. The same normalization value is chosen for the t-y planes of the same volunteer. Learning is a procedure that adjusts weights of neural network so that the output is close to the target for the input. A convolutional neural network (CNN), known as U-net, is constructed for compressed data with CRs of 2, 3, and 4, as shown in Fig.2. Since the input to the t-y plane neural network is not square matrices (20 x 256), the sizes of max-pooling and transpose convolution vary from layer to layer. Reconstruction for compressed sensing CINE MRI using the t-y plane is shown in Fig.3. The ‘initial reconstruction CINE’ images are sliced into t-y planes, and then fed to the neural network as input after normalization. The network output is repeatedly denormalized and stored for the entire x to create estimated ‘error CINE’ images. The error CINE images are added to the initial reconstruction for final reconstruction.Results
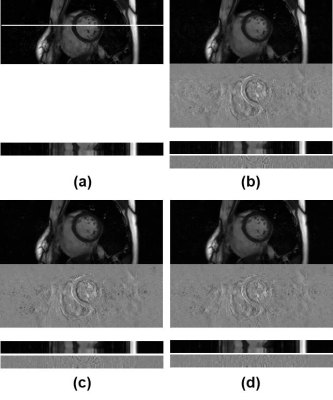
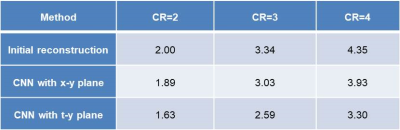
Figure 4 shows test images reconstructed with (a) ground-truth imaging, (b) initial reconstruction, (c) neural network using x-y plane, and (d) neural network using t-y plane. The upper images show transverse planes, and the lower images stacked line profiles along cardiac phase vertically. Difference images between the reconstructed images and the ground-truth images are also shown below. The average normalized mean square error for the reconstructed images are summarized in Table 1 for the test data. As shown in Fig.4 and Table 1, neural network improves reconstruction substantially compared to initial reconstruction. The neural network based on the t-y plane shows better cross-sectional views, clearer temporal profiles, and less normalized mean square errors compared to that of the x-y plane.Conclusion
We build a deep neural network in time-phase encoding plane (t-y) for compressed sensing cardiovascular CINE MRI. By adopting the t-y plane, instead of the x-y plane, simultaneous restoration in time and space is effectively achieved. For test data, neural network based on the t-y plane shows the best performance.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2019R1A2C2005660). The present research has also been conducted by the research grant of Kwangwoon University in 2019.References
[1] Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, Cham. 2015; p. 234-241.
[2] Yoon JH, Kim PK, Yang YJ, Park J, Choi BW, Ahn CB. Biases in the Assessment of Left Ventricular function by Compressed Sensing Cardiovascular Cine MRI. Investigative Magnetic Resonance Imaging. 2019 Jun;23(2):114-124.
[3] Park J, Hong HJ, Yang YJ, Ahn CB. Fast cardiac CINE MRI by iterative truncation of small transformed coefficients. Investigative Magnetic Resonance Imaging. 2015 Jan;19(1):19-30.
Figures