3373
Relaxation-Time Selective Imaging Using the Inverse Z-transform1Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of, 2Division of Biomedical Engineering, Hankuk University of Foregn Studies, Yongin, Korea, Republic of, 3Imaing Institute, Cleveland Clinic Foundation, Cleveland, OH, United States, 4Center for Neuroscience Imaging Research, Suwon, Korea, Republic of
Synopsis
There have been many attempts to separate mixed relaxation-time decays. Among them, some approaches do not assume the number of exponential decays a priori for the analysis of multiple-component decay signals. However, they are sensitive to ill conditions and have a poor resolving ability in terms of decay constants. In this study, a new method is proposed that can analyze multiple T2 decays with high resolution using the inverse Z-transform, which was demonstrated in simulation and in vivo human brain experiment. T2-selective images were also presented and used for myelin-water fraction mapping and deep brain-tissue segmentation.
Purpose
Signals with multiple exponential decays are often encountered in science and engineering. In MR field, T1 and T2 relaxation times is included. In many cases, we acquire MR signals which contain multiple T2 components in a single voxel. There are previous attempts to decompose multiple-exponential decays without assuming the number of decays a priori such as using the inverse Laplace transform 1-2 or linear optimization methods.3-5 However, they are sensitive to ill conditions and have a poor resolving. In this study, we propose a new method that can analyze multiple exponential decays like T2 decays with high resolution using the inverse Z-transform. It was demonstrated in simulation and in vivo human brain experiment, providing T2-selective images and using them for myelin-water fraction mapping and deep brain-tissue segmentation.Methods
Theory: The rationale for using the inverse Laplace transform (L-1) originally comes from L-1{exp(-snT)} = δ(t-nT), where T is a sampling interval in t-domain and n is a discrete sampling index. For applying the Z-transform, z can be defined as zk = exp(skT): L-1(zk-n) = δ(t-nT), where k is an integer from 0 to M-1 (M is a given integer). Therefore, it should be noted that, during the following discussion, acquired MRI signals belong to the s-domain, NOT to the t-domain. Assuming that there is a finite and discrete function, g(t), in t-domain, theng(t) = ΣNn=1 xn δ(t-nT) [1]
L{g(t)} = L{ΣNn=1 xn δ(t-nT)} = ΣNn=1 xn exp(-snT) [2]
= ΣNn=1 xn z-n = Z(g(t)) [3]
Eq.[2] and [3] show that a sum of T2 exponential decays can be transformed to g(t), the sum of δ functions, using the inverse Z-transform. Since z = exp(skT), z can be redefined as AWk (Fig. 1), where A = A0exp(jθ0) and W = W0exp(jφ0), θ0 and φ0 are the initial angles of A and W, decided arbitrarily.6 Then we can rewrite Eq.[3] in form of column vectors (X = [Xk], length M and x = [xn], length N) and a coefficient matrix (C = [(AWk)n]):
X = Cx [4]
The final results we want, i.e., the decomposed multi-component exponential decays, are the components of x, which is given by x = C-1X (M = N). Components of x indicate which T2 decays are contained in the acquired MR signal, X.
Simulation: To test the effect of the inverse Z-transform in simulation, we built a synthetic phantom in MATLAB containing four different exponential decays with different R2(=1/T2)[1/ms] and amplitudes (Fig. 2A): 1000×exp(-0.046t), 3600×exp(-0.032t), 7500×exp(-0.02t), and 2000×exp(-0.01t), with 3% random noises. Both the first TE and ΔTE were assumed to be 10 ms, with 30 sample points. If the inverse Z-transform is working properly, it is expected that each exponential decay is separately displayed with the estimated R2 values and magnitudes.
Human brain Experiment: Human brain experiment was also performed in the Siemens 3T Prisma system not only to test the proposed method itself, but also to explore some promising applications. A multi-echo GRASE sequence was used to obtain the myelin-water fraction(MWF) maps for reference. 40 slices were acquired and, for each slice, 32 echo images were acquired at varying TE from 10 to 320 ms by an increment of 10 ms. Scan parameters are given in Fig.4.
Results and Discussion
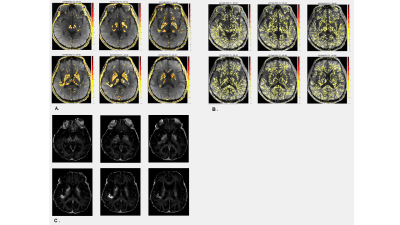
Figures 2B~E shows the R2-selected images of the synthetic phantom, each of which corresponds to four components, respectively. Each exponential decay was successfully selected and displayed. The estimated amplitudes of each exponential decay across all the voxels in each T2-selected image were: 999.94±0.21(B), 2000.05±0.31(C), 3599.80±0.49(D), 7500.10±0.30(E), respectively, and the estimated R2 values were: 0.045±0.007(B), 0.010±0.014(C), 0.033±0.007(D), 0.019±0.008(E). They were in excellent agreement with the original amplitudes and R2 values.Figure 3 shows T2-selective human brain images (A, B) and MWF maps obtained from multi-echo GRASE for reference (C). Two T2 ranges were selected by the proposed method, that is, 45~55 ms(A) and 55~65 ms(B), respectively. It is interesting that the shorter T2-selective images (A) seem to separate deep brain structures such as thalamus (arrow 1), putamen and globus pallidus (arrow 2), whereas the longer T2-selective images (B) seem to represent the MWF maps except for the deep brain structures shown in Fig. 3A. A combination of Figs. 3A and B looks like the actual MWF maps, which are good agreement with Fig. 3C.
Conclusion
In this study, we proposed a method that effectively decomposes the relaxation-time decays, e.g., T2 decays, using the inverse Z-transform. The method can be easily applied to T1 decays. Its performance was successfully demonstrated in simulation and in vivo human brain experiment. The proposed method can provide T2-selective imaging with T2 ranges what we want. In addition, when compared to the previous works using the inverse Laplace transform 1,2 or a linear inversion program 4,5, it is less sensitive to ill-posed problems and provide highly T2-resolved images. It is also expected to find uses in many interesting applications such as MWF mapping and accurate segmentation of brain tissue including subcortical gray matters etc. A further study is warranted for investigating the maximum T2 range that can be resolved, the highest T2 resolution that can be achieved, and the relationship between T2 range and T2 resolution.Acknowledgements
This work was supported by SMC-SKKU Future Convergence Technology Program.References
1. Labodie, C et al. Relaxographic imaging. J Magn Reson B105, 99-112 (1994)
2. Fenrich, FRE et al. Relaxation times and microstructures, NMR Biomed 14, 133-139 (2001)
3. Kroeker, RM & Kenkelman, RM. Analysis of biological NMR relaxation data with continuous distributions of relaxation times. J Magn Reson 69, 218-235 (1986)
4. Provencher, SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput Phys Commun 27, 213-228 (1982)
5. Provencher, SW. CONTIN. A general purpose constrained regularization program for inverting noisy linear algebraic or integral equations. Comput Phys Commun 27, 229-242 (1982)
6. Rabiner, LR. The Chirp Z transform algorithm. IEEE Trans Audio Electroacoust 17, 87-92 (1969).
Figures