3370
Evaluation of Deep Learning Techniques for Motion Artifacts Removal
Alessandro Sciarra1,2, Soumick Chatterjee2,3, Max Dünnwald1,4, Oliver Speck2,5,6,7, and Steffen Oeltze-Jafra1,5
1MedDigit, Department of Neurology, Medical Faculty, Otto von Guericke University, Magdeburg, Germany, 2BMMR, Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 3Data & Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 4Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 5Center for Behavioral Brain Sciences (CBBS), Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
1MedDigit, Department of Neurology, Medical Faculty, Otto von Guericke University, Magdeburg, Germany, 2BMMR, Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 3Data & Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 4Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 5Center for Behavioral Brain Sciences (CBBS), Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
Synopsis
Removing motion artifacts in MR images remains a challenging task. In this work, we employed 2 convolutional neural networks, a conditional generative adversarial network (c-GAN), also known as pix2pix, as well as a network based on the residual network (ResNet) architecture, to remove synthetic motion artifacts for phantom images and T1-w brain images. The corrected images were compared with the ground-truth ones in order to assess the performance of the chosen neural networks quantitatively and qualitatively.
Introduction
Motion artifacts are still an open problem in research and clinical routine MR acquisitions1. Despite the efforts to solve or limit the degradation effects on images caused by motion, corrupted images with relatively poor image quality are no exception. In addition to the traditional motion correction techniques, such as prospective motion correction, retrospective motion correction, etc.2, the number of attempts that use deep learning techniques in order to remove motion artifacts increased in recent years3-6. In this work, we comparatively evaluated the performance of two neural networks for the task of motion artifacts removal.Methods
Two datasets were compiled using first, the T1-weighted (T1-w) images from the IXI clinical neuroimaging repository7 and second, the 2D Shepp-Logan phantom. Moreover, two neural networks were utilized: Pix2pix by Philip Isola et al.8, a conditional adversarial network, originally proposed for Image-to-Image Translation and a ResNet-based network9, originally proposed for undersampled MRI reconstruction. Motion artifacts were artificially created using a custom-made Python function, which randomly modifies the corresponding k-space images and then, converts them back to the image space (left column in Fig. 1 and 2). Consequently, the level of motion corruption was also random, but classified into three main categories: low, mild and heavy level of corruption (left column in Fig. 2). The pix2pix network was configured with a U-Net10 generator and a PatchGAN8 discriminator, implemented in TensorFlow by Yen-Chen Lin11. ResNet12 was initially proposed for image recognition. Our residual network9 uses a modified version of the Residual Block, by adding a Spatial Dropout13, which randomly drops or zeros out feature maps, between the two convolution layers of the Residual Block. Pytorch was used to implement this network. For Pix2Pix, the dataset contained 2000/1000/1000 paired samples (corrupted & ground truth), for training, validation, and testing, respectively. For ResNet, the dataset contained only the central slices of each volume, from the 60th to the 90th slice, of 100/35/50 subjects (3100/1085/1550 images), for training, validation and testing, respectively. For initial tests using ResNet (ResNet-1), one random angle was chosen for each slice, and all the lines of that particular slice were rotated using that same angle. For the second test using ResNet (ResNet-2) and for the pix2pix test, images were synthetically corrupted in the same manner.The obtained results were analyzed by calculating the mean-squared-error (MSE). Furthermore, after skull-stripping and three tissues segmentation (gray matter, GM, white matter, WM and cerebrospinal fluid, CSF) performed using FSL14, a multi-class DICE coefficient15 and volume ratios were calculated. The volume ratio is computed by dividing the number of pixels of each tissue class, e.g., the number of pixels for GM in the predicted image, by the number of pixels of the same class in the ground-truth image. The processing pipeline is shown in Figure 3.Results
Figure 1 shows two predictions for the Shepp-Logan phantom. The prediction has been tested without (upper row) and with (lower row) previously adding field inhomogeneities to the phantom images. In Figure 2, motion artifacts removal in T1-w images using the pix2pix network is illustrated. In Figure 4, three box plots show the MSE values, DICE coefficients and the volume ratios, respectively (from left to right). The MSE values, are calculated first, between input (corrupted image) and ground-truth (free-motion image) and second, between the predicted/corrected and the ground-truth image. The DICE coefficients are computed in a similar fashion. In this case, the values are the averages across the three considered classes GM, WM and CSF. The last box plot shows the volume ratios, also, averaged across the three classes. For ResNet, two results corresponding to the two different difficulty levels (ResNet-1 and ResNet-2) are illustrated.Discussion
Removing motion artifacts from 2D-Shepp-Logan phantom images seems to work reliably using pix2pix. However, it is possible that the network learnt the shape of the phantom and consequently, extracted it from the corrupted images (Fig. 1). For the MR brain images, both networks correctly removed the motion artifacts, but could not consistently reconstruct the images with all of their details. MSE values decrease after the application of the neural nets. This is in agreement with the visual comparison, where most of the motion artifacts are removed from the corrupted images, but again, in case of a heavy corruption level, the network is not able to retrieve back all the edge information and the small structures are not visible. The DICE coefficients and the volume ratios, on the other hand, indicate a good agreement between the segmentation of the corrected and the ground-truth images. ResNet-1 provides the best results with the lowest MSE values, the highest DICE coefficient, and almost 0.8 and 1.0 mean value for the volume ratio. After increasing the difficulty level, the result slightly deteriorates, but still, ResNet-2 outperforms pix2pix.Conclusion
Qualitative and quantitative assessments showed that the chosen networks are able to remove motion artifacts up to a certain level of corruption. While the results are encouraging, it remains a challenge to determine good hyperparameters of the networks and their training. In a comparison of ResNet and pix2pix, ResNet outperformed pix2pix.Acknowledgements
This work was supported by the federal state of Saxony-Anhalt under grant number ‘I 88’ (MedDigit); and was in part conducted within the context of the International Graduate School Memorial at OvGU (project no. ZS/2016/08/80646).References
- Zaitsev, M. et al.: Motion Artefacts in MRI: a Complex Problem with Many Partial Solutions. J MagnReson Imaging. 2015 Oct; 42(4):887-901. doi:10.1002/jmri.24850
- Godenschweger, F. et al: Motion correction in MRI of the brain. Phys. Med. Biol. 61 (2016) R32–R56. doi:10.1088/0031-9155/61/5/R32
- Küstner, T. et al.:Retrospective correction of motion‐affected MR images using deep learning frameworks. Magn Reson Med. 2019;82:1527–1540. DOI: 10.1002/mrm.27783
- Jiang, W. et al.: Respiratory Motion Correction in Abdominal MRI using a Densely Connected U-Net with GAN-guided Training. https://arxiv.org/ftp/arxiv/papers/1906/1906.09745.pdf
- Oksuz, I. et al.: Cardiac MR Motion Artefact Correction from K-space Using Deep Learning-Based Reconstruction. MLMIR 2018: Machine Learning for Medical Image Reconstruction pp 21-29. https://doi.org/10.1007/978-3-030-00129-2_3
- Usman, M. et al.: Motion Corrected Multishot MRI Reconstruction Using Generative Networks with Sensitivity Encoding. https://arxiv.org/pdf/1902.07430.pdf
- IXI dataset: https://brain-development.org/ixi-dataset/
- Phillip Isola, Jun-Yan Zhu, Tinghui Zhou, Alexei A. Efros. “Image-to-Image Translation with Conditional Adversarial Networks”. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). DOI: 10.1109/CVPR.2017.632, https://arxiv.org/pdf/1611.07004v1.pdf
- Chatterjee, S. et. al.: A deep learning approach for reconstruction of undersampled Cartesian and Radial data. ESMRMB 2019
- Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
- Pix2pix tensor-flow implementation: https://github.com/yenchenlin/pix2pix-tensorflow
- He K. et. al: Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). DOI: https://doi.org/10.1109/CVPR.2016.90
- Tompson, J., Goroshin, R., Jain, A., LeCun, Y., Bregler, C.: Efficient object localization using convolutional networks, http://arxiv.org/pdf/1411.4280v3
- FSL: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki
- Dice, L. R.: Measures of the Amount of Ecologic Association Between Species. Ecology 1945; 26(3):297-302.
Figures

Motion correction using Pix2Pix for Sheep-Logan phantom. Upper row: Input) Sheep-Logan phantom corrupted with artificial motion artifacts, Predicted) image obtained from Pix2Pix, Ground-truth) original image without motion artifacts. Bottom row: similar to the upper row, but with additional inhomogeneities.
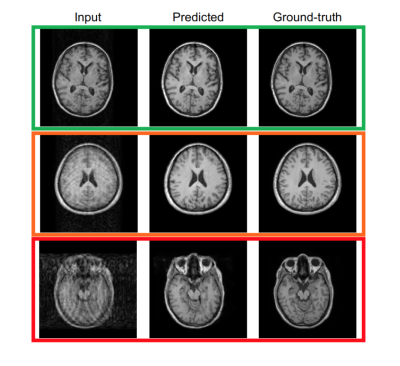
Classification into three main categories/scenarios. Upper row: input image slightly corrupted by motion artifacts. Middle row: input image with medium level of corruption. Bottom row: input image heavily corrupted by motion artifacts, even large structures are not easy to recognize.

Pipeline used for processing the results.

Results: Mean-squared-error (MSE), DICE coefficient and Volume ratios. The green triangles indicate the mean values and the lines the correspondent median values