3359
Motion Correction Strategy for Multi-Contrast based 3D parametric imaging: Application to Inhomogeneous Magnetization Transfer (ihMT)1Aix-Marseille Univ, CNRS, CRMBM, Marseille, France, 2APHM, Hôpital Universitaire Timone, CEMEREM, Marseille, France, 3Université de Strasbourg, CNRS, ICube, FMTS, Strasbourg, France, 4APHM, Hôpital Universitaire Timone, Service de neurologie, Marseille, France, 5Division of MR Research, Radiology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, United States
Synopsis
Multi-contrast-based parametric MRI techniques provide parametric maps intrinsically sensitive to voxel misalignment upon images combination.
In this work, we propose an efficient retrospective motion correction method adapted to this problem, with an application to inhomogeneous Magnetization Transfer (ihMT) imaging. The proposed method is confronted with the MCFLIRT (FSL) method used as reference, and residual motion is quantified and analysed across native and motion corrected images.
Introduction
Parametric images based on the combination of MRI images with varying contrasts for composite map generation, such as inhomogeneous Magnetization Transfer (ihMT), are of great interest for structural evaluations in the central nervous system1,2. However, potential voxels misalignment across acquired images, induced for instance by subject motion, may result in inconsistencies in the computed maps (loss of sharpness or localized artifacts), which bias further analyses. Compensation schemes to account for images misalignment must thus be considered.In this work, we propose an efficient and robust retrospective motion correction framework for multi-contrast-based imaging techniques (MC-MoCo) accounting for a limited number of available images as specific feature. The proposed method was applied on 3D ihMT images acquired with a clinical routine procedure3 on healthy subjects and Multiple Sclerosis (MS) patients. The MC-MoCo method was compared to MCFLIRT4, available in the FSL package, and showed higher performances for motion correction for both populations.
Materials and methods
Imaging experiments were performed at 1.5T (Avanto, Siemens, Erlangen, Germany) using a sensitivity-enhanced 3D ihMT-GRE3 sequence on 6 healthy volunteers and 7 patients with relapsing-remitting MS.Thirteen 3D images were sequentially acquired and consisted on a reference volume (MT0, no radiofrequency saturation prior to image acquisition), followed by three repetitions of 4 different frequency-offset (Δf) MT-weighted (MTw) volumes (MT+ (Δf=+8 kHz), MT+- (Δf=+/-8 kHz), MT- (Δf=-8 kHz) and MT-+ (Δf=-/+8 kHz))3.
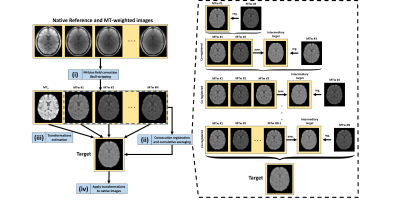
Figure 1 illustrates the MC-MoCo method. The whole process, which relies on functionalities provided in the ANTs registration package5, can be summed up as follows: (i) N4 bias-field correction6 and template-based skull-stripping of the native volumes, (ii) target building based of the MTw images, (iii) registration of each native skull-stripped volumes onto the target, (iv) application of the estimated transformations to each native volume. The MC-MoCo method was compared to MCFLIRT4 using the averaging option for target building in order to avoid the bias associated to the target choice among the various available contrasts in the case of a single available target option. Both MC-MoCo and MCFLIRT methods exclusively utilized rigid-body transformations and the mutual information metric7 to account for the various contrasts.
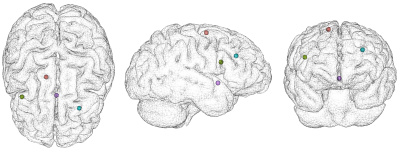
Residual motion following the application of motion correction methods was used to evaluate the efficiency of MC-MoCo and MCFLIRT. For that, landmarks were positioned over each native and motion-corrected volume at specific anatomical regions depicted in Figure 2, using 3D Slicer8. For each image group (uncorrected, MCFLIRT, and MC-MoCo), each subject and each marker, the barycentre of the respective locations on every volume was computed to serve as a reference point for the estimation of the residual movement. This metric, defined as the mean distance to the barycentre over every volume, quantifies the efficiency of a motion correction method for a single marker and a single subject: more efficient methods will yield lower residual motion.
Statistical analyses were performed to test for the significance of MC-MoCo and MCFLIRT on correcting motion and to ensure that both methods performed similarly for each marker and each subject group (control or MS); we hence estimated a mixed-effects linear model, with fixed effects on the motion correction method, the marker, and the subject group, and with a random effect on the subject, added to account for the non-independence of the observations within a subject.
Results
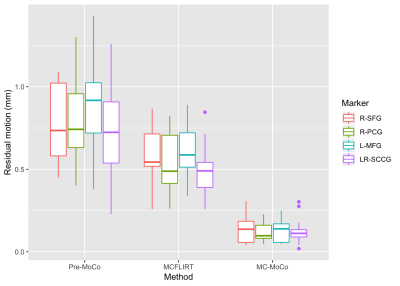
A boxplot of the residual motion grouped before and after corrections is shown in Figure 3: this plot indicates that MC-MoCo has less residual motion than MCFLIRT and that this behavior is rather constant across all markers.The ANOVA on the estimated model shows a significant effect of the motion-correction method (p<0.01), but no significant effect of the marker (p=0.14) or of the group of the subject (p=0.12). Post-hoc tests, with p-values corrected for multiple comparison, show that residual motion is significantly reduced with respect to the non-corrected images by both MCFLIRT (-0.43 mm, p<0.01) and MC-MoCo (-0.68 mm, p<0.01), and that MC-MoCo significantly outperforms MCFLIRT (p<0.01). Both MC-MoCo and MCFLIRT performed similarly for controls and MS patients (no significant group effect, p>0.05).
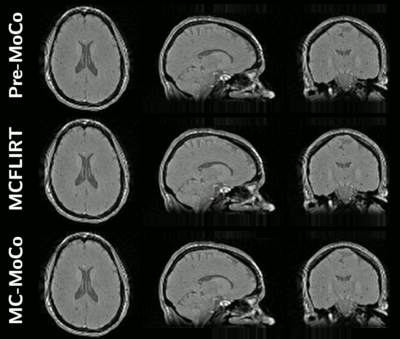
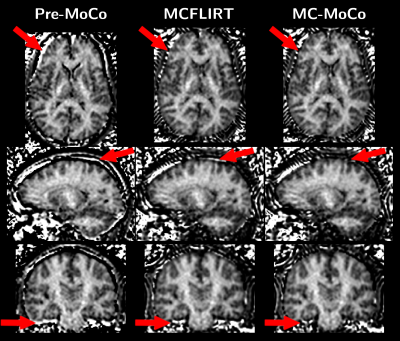
Representative animated MT0 and MTw images in orthogonal views for one subject before and after motion corrections are provided in Figure 4. Figure 5 shows representative ihMT images in orthogonal views, depicting the removal of artifacts relative to misalignment correction when using MCFLIRT and MC-MoCo.
Discussion and conclusion
The key point of the MC-MoCo method lies in the target building step. As opposed to other motion-correction methods based either on an averaged target over all native images or a single selected reference volume, the target is built using an iterative and updated pattern, therefore providing a reliable result given a limited number of images. The MC-MoCo method applied in the context of 3D ihMT images significantly decreased the residual motion and outperforms MCFLIRT.Acknowledgements
This work was supported by the SATT Sud-Est (France), the French Association pour la Recherche sur la Sclérose En Plaques (ARSEP), Roche Research Foundation (Switzerland) and French National Research Agency, ANR [ANR‐17‐CE18‐0030]. This work was performed by a laboratory member of France Life Imaging network (grant ANR-11-INBS-0006).
The authors thank V. Gimenez for management.
References
1. Duhamel G, Prevost VH, Cayre M, et al. Validating the sensitivity of inhomogeneous magnetization transfer (ihMT) MRI to myelin with fluorescence microscopy. Neuroimage 2019;199:289–303 doi: 10.1016/j.neuroimage.2019.05.061 SMASH.
2. Varma G, Duhamel G, de Bazelaire C, Alsop DC. Magnetization transfer from inhomogeneously broadened lines: A potential marker for myelin. Magn. Reson. Med. 2015;73:614–622 doi: 10.1002/mrm.25174 SMASH.
3. Mchinda S, Varma G, Prevost VH, et al. Whole brain inhomogeneous magnetization transfer (ihMT) imaging: Sensitivity enhancement within a steady-state gradient echo sequence. Magn. Reson. Med. 2018;79:2607–2619 doi: 10.1002/mrm.26907 SMASH.
4. Jenkinson M, Bannister P, Brady M, Smith S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. Neuroimage 2002;17:825–841 doi: 10.1006/nimg.2002.1132 SMASH.
5. Avants BB, Tustison N, Song G. Advanced Normalization Tools (ANTS). Insight J. 2 2009:1–35.
6. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging 2010;29:1310–1320 doi: 10.1109/TMI.2010.2046908 SMASH.
7. Viola P, Wells III WM. Alignment by Maximization of Mutual Information. Int. J. Comput. Vis. 1997;24:137–154 doi: 10.1023/A:1007958904918 SMASH.
8. Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012;30:1323–1341 doi: 10.1016/j.mri.2012.05.001 SMASH.
Figures