3358
Clinical Utility of Deep Learning Motion Correction for Neuroimaging
Kamlesh Pawar1, Jarrel Seah2, Meng Law3, Tom Close4, Zhaolin Chen1, and Gary Egan1
1Monash Biomedical Imaging, Monash University, Melbourne, Australia, 2Department of Neuroscience, Monash University, Melbourne, Australia, 3Radiology and Nuclear Medicine, Alfred Health, Melbourne, Australia, 4Monash Biomedical Imging, Monash University, Melbourne, Australia
1Monash Biomedical Imaging, Monash University, Melbourne, Australia, 2Department of Neuroscience, Monash University, Melbourne, Australia, 3Radiology and Nuclear Medicine, Alfred Health, Melbourne, Australia, 4Monash Biomedical Imging, Monash University, Melbourne, Australia
Synopsis
The deep learning techniques have been shown to reduce the motion artifact in simulated motion scenarios and a few volunteer scans, the validation of it during routine clinical scans remains an unanswered question. In this study, we focus on evaluating the quality of images from the DL motion correction approach on a cohort of 27 actual patient motion cases that were obtained from the routine clinical scans. Two board-certified radiologists evaluated 9 anatomical regions in the 3D MPRAGE brain images and rated them on the 3-point scale.
Introduction
Patient motion remains a major cause of artifact in MR imaging, particularly in 3D scans. Various methods for minimizing the effect of motion have been proposed including navigator1, the external camera2 and data-driven approaches3-7. Each approach has its advantages and limitations. For instance, navigators can provide an accurate estimate of motion, however, they come at the cost of increased scan time in some sequences, and it may not be possible to integrate them in other sequences. Camera-based systems can track head motion accurately but their performance may be degraded by occlusion of the head by the receive coils, which makes camera-based solutions complex to set up and calibrate. Deep learning motion correction is a data-driven approach that does not have the aforementioned limitations. Although DL techniques have been shown to reduce motion artifact in simulated motion scenarios and a few volunteer scans, their application to routine clinical scans has not been validated. In this study, we evalulate DL motion correction on 27 routine clinical scans that were corrupted by motion. Two board-certified radiologists evaluated 9 anatomical regions in the 3D MPRAGE brain images and rated them on the 3-point scale.Methods
Deep Learning Network DesignAn encoder-decoder7 image to image translation deep learning network shown in Figure. 1 was designed to suppress motion artifact from 3D MPRAGE images. The encoder consisted of the InceptionResNet-V2 architecture and the decoder consisted of a series of convolution and upsampling layers. For the layer that has equal spatial dimensions in the encoder and decoder, were concatenated (skip connections) in order to pass the spatial information across. The network was trained with a pair of motion degraded and motion-free images. The motion degraded images were generated with a simulation that takes a clean image and motion parameters (3 rotation and 3 translation) as input and provides corresponding motion degraded images. We created 1572 3D MPARGE simulated motion datasets for training and 384 motion datasets for validation of the network using the IXI dataset (https://brain-development.org/ixi-dataset/).
Image Quality Assessment
The trained network was used to correct the motion artifact from 27 real motion degraded cases from routine clinical scans. The study was approved by the Hospital’s ethics/IRB committee and two board certificated radiologists evaluated the image quality of both the motion degraded and motion-corrected scans for 9 anatomical brain regions. The anatomical regions were: ventricles, grey-white junction, subarachnoid space sharpness, brainstem, cerebellar, basal ganglia, circle of Willis, fornix, and hippocampus. The scoring was based on a 3-point scale: 3 = well-defined structure, 2 = structure seen but not well defined and 1 = structure not seen.
Results and Discussion
The quantitative assessment of image quality scores showed marked improvements in the perceptual image quality as demonstrated from the bar chart in Figure 2. Paired t-tests across all the evaluated regions were significant (p< 0.005), demonstrating that there was a consistent improvement in the image quality using the deep learning motion correction method. The hippocampus was the region that benefited most from the proposed motion correction technique, where the radiologist score improved by 0.82 on a 3-point scale. The ventricles showed the least improvement of 0.32 on a 3-point scale. The ventricles did not show a great deal of improvement because the region was also least affected by patient motion. In general, the larger regions (ventricles) were least affected by the motion and smaller regions such as the hippocampus were most affected by the motion. The mean score after the motion correction was at least 2.41 (circle of Willis) suggest that all the structures were at least visible seen after the motion correction. Three orthogonal slices of a representative case are shown in Figure 3 where the green arrow indicates the hippocampus (panel-a), brainstem (panel-b) and fornix (panel-c) respectively. Reduced ghosting, ringing and enhanced delineation (red arrow) of grey-white matter can be observed across the three orthogonal views. The proposed DL motion correction technique is a post-processing technique and thus can be seamlessly integrated into the clinical imaging pipeline.Conclusion
The clinical evaluation of DL motion correction of 27 real patient motion cases demonstrates that the proposed technique improves the perceptual image quality of MR images and able to improve the visibility of all the brain regions. The plug-and-play nature of the method makes it readily available for routine clinical application.Acknowledgements
No acknowledgement found.References
- Fu ZW, Wang Y, Grimm RC, Rossman PJ, Felmlee JP, Riederer SJ, Ehman RL. Orbital navigator echoes for motion measurements in magnetic resonance imaging. Magnetic resonance in medicine. 1995 Nov;34(5):746-53.
- Zaitsev M, Dold C, Sakas G, Hennig J, Speck O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage. 2006 Jul 1;31(3):1038-50.
- Loktyushin A, Nickisch H, Pohmann R, Schölkopf B. Blind retrospective motion correction of MR images. Magnetic resonance in medicine. 2013 Dec;70(6):1608-18.
- Küstner T, Armanious K, Yang J, Yang B, Schick F, Gatidis S. Retrospective correction of motion‐affected MR images using deep learning frameworks. Magnetic resonance in medicine. 2019 May 13.
- Johnson PM, Drangova M. Conditional generative adversarial network for 3D rigid‐body motion correction in MRI. Magnetic resonance in medicine. 2019 Sep;82(3):901-10.
- Pawar K, Chen Z, Shah NJ, Egan GF. Motion correction in MRI using deep convolutional neural network. In Proceedings of the ISMRM Scientific Meeting & Exhibition, Paris 2018 Jun (Vol. 1174).
- Pawar K, Chen Z, Shah NJ, Egan GF. Suppressing Motion Artefacts in MRI using an Inception-Resnet Network with Motion Simulation Augmentation. NMR in Biomedicine; 2019 Oct 24. (in press), doi: 10.1002/nbm.4225.
Figures
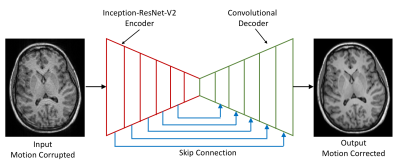
Figure 1: Deep learning image to image translation architecture for motion correction; the encoder (red) consisted of Inception-ResNet-V2 architecture and the decoder (green) consisted of a series of convolution and upsampling layers; skip connection (blue) were made in the form of concatenation from encoder to decoder to reinsert the spatial resolution.
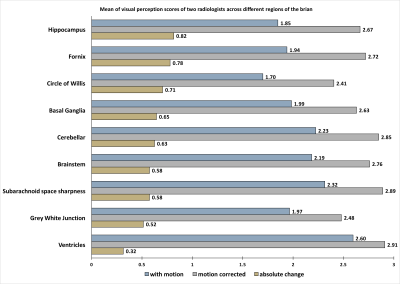
Figure 2: Mean of visual perception of two radiologist across different regions of the brain on a 3-point scale: 3 = well defined structure, 2 = structure seen but not well defined and 1 = structure not seen. The highest visual perception improvement was observed in hippocampus (0.82 points) while the lowest improvement was observed in ventricles (0.32 points). Although the ventricles were least improved but were also least affected by the motion as observed from the score of 2.6 for motion corrupted images.
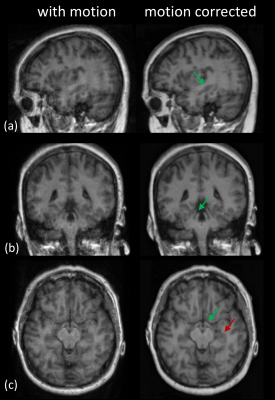
Figure 3: A representative image showing the effect motion correction; (a): green arrow showing that the visibility of the hippocampus improved after motion correction; (b): green arrow shows the brainstem which was clearly delineated after motion correction; (c): green arrow clearly showing of the fornix which was blurred due to patient motion, red arrow showing the clear demarcation of grey-white boundary.