3350
Non-rigid Respiratory Motion Estimation of Coronary MR Angiography using Unsupervised Fully Convolutional Neural Network1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom
Synopsis
Non-rigid motion corrected coronary MR angiography (CMRA) in combination with 2D image-based navigators has been proposed to account for the complex respiratory-induced motion of the heart in undersampled acquisitions. However, this framework requires the efficient and accurate estimation of non-rigid bin-to-bin motion from undersampled respiratory-resolved images. In this study, we aim to investigate the feasibility of using an unsupervised fully convolutional network to estimate non-rigid motion from undersampled respiratory-resolved CMRA. The performance of the proposed approach was evaluated on 5-fold accelerated free-breathing CMRA and validated against a widely used conventional non-rigid registration method.
Introduction
Respiratory motion compensation is essential to enable free-breathing whole-heart coronary MR angiography (CMRA) with 100% scan efficiency. Recently, non-rigid motion corrected CMRA (1-3) in combination with 2D image-based navigators (iNAVs) (4) has been proposed to account for the complex respiratory-induced motion of the heart in undersampled acquisitions. This approach bins the data in different respiratory positions based on iNAVs, and corrects translational intra-bin motion (estimated from iNAVs) and non-rigid inter-bin motion (estimated from undersampled respiratory-resolved bin images). Therefore, it requires efficient and accurate estimation of non-rigid respiratory motion fields, which is challenging and computationally demanding. Convolutional neural networks have been recently proposed to estimate dense optical flow of natural images and achieved promising results (5,6). However, these approaches have not been extended to estimate non-rigid motion from undersampled MR images, which is challenging due to no gourd truth for supervised training, fine motion to be estimated and contrast changes between the motion frames. In this study, we proposed a novel fully convolutional network trained in an unsupervised fashion (UFCN) to estimate non-rigid bin-to-bin respiratory motion.Methods
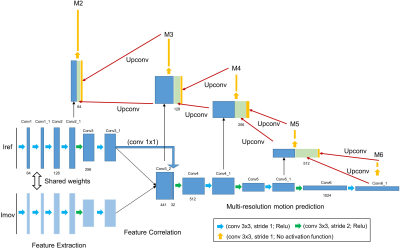
Motion estimation Network: The proposed motion estimation network (Fig.1) takes a pair of images, Iref (end-expiration reference bin) and Imov (one of the other respiratory bins) as input, and outputs the non-rigid motion fields from Iref to Imov. The network includes 3 parts: feature extraction; correlation of features from Iref and Imov; multi-scale registration which estimates motion at 16x, 8x, 4x, 2x downsampled resolution and full resolution (M6-M2 in Fig. 1).End-to-end unsupervised learning: Due to the difficulty of obtaining ground truth non-rigid motion fields in real acquisitions, the network is trained in an unsupervised way. The basis of the unsupervised learning is that each pixel $$$x$$$ in Imov warped by motion field $$$M_{x}$$$ should be similar to the pixel $$$x$$$ in Iref, which can be promoted via data loss. Additionally, the motion fields should be smooth, which can be encouraged with smoothness constraint. Therefore, the unsupervised loss to minimize is:$$1-NCC\left(I_{ref}\left(x\right),I_{mov}\left(M_{x}+x\right)\right)+\lambda R\left(M_{x}\right) [1]$$
where NCC computes the normalized cross correlation between Iref and Imov warped with estimated motion field $$$M_{x}$$$ and is used to encourage the similarity between Iref and warped Imov. $$$R\left(M_{x}\right)$$$ is a second-order smoothness constraint on the motion fields that is used to encourage collinearity of neighbouring motion pixels (4). $$$\lambda$$$ is the weight of the regularization term. Each motion prediction has the loss terms defined in Eq. [1], and the total loss is the weighted summation of losses for all five multi-scale motion fields (M6-M2 in Fig. 1).
Data: Whole-heart free-breathing CMRA was acquired as described in (3) with 5-fold acceleration and 1.2mm isotropic spatial resolution on 17 healthy subjects. The undersampled CMRA data was sorted into 3 respiratory bins based on beat-to-beat foot-head translational motion estimated from iNAVs. Intra-bin translational motion correction was performed for each bin and respiratory bin images (~15-fold accelerated) were reconstructed with soft-gating iterative SENSE to reduce undersampling artifacts. For each subject, forty 2D slices covering the heart were considered, resulting in 1360 samples with each sample defined as a pair of images at end-expiration (Iref) and at one of the other respiration states (Imov). 95% (1292) of the samples were randomly selected as training data, whereas the remaining 68 samples were used for testing.
Network training: The network was trained for 20000 iterations. For each iteration, 10 samples were randomly selected to constitute one batch. Data augmentation included random horizontal flipping and affine transformation. The initial learning rate was set at 0.0001 and reduced by half every 5000 iterations. To reduce overfitting, l2-norm regularization of the network weights was imposed.
Evaluation: Once trained, the proposed UFCN was validated against a state-of-art conventional multi-scale registration method (7) in NIFTI registration toolbox, which also uses NCC as registration metric and constrained the motion field smoothness. All parameters of NIFTI registration were carefully optimized for the specific application. Registration performance was evaluated by calculating the NCC between Iref and Imov, and between Iref and warped Imov using the corresponding estimated motion.
Results
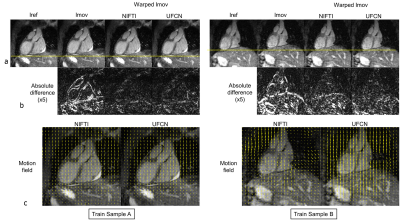
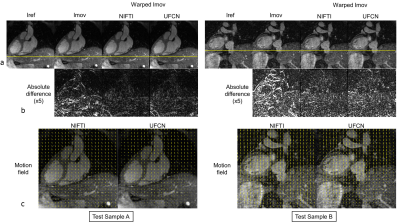
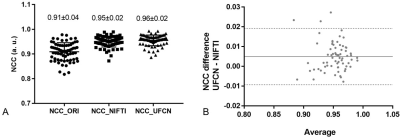
Network training took about 5 hours. Once trained, UFCN estimated the motion fields in 0.28±0.01s per sample, 50 times faster than NIFTI (14.52±2.57s). Representative motion estimation results of training and test samples are shown in Fig. 2 and Fig. 3, respectively. Overall, the proposed UFCN achieved similar motion estimation performance to NIFTI in terms of absolute difference and NCC, whereas better motion estimation was given by UFCN for some samples as one example shown in Train Sample B in Fig. 2. For the 68 test samples, NCCs before registration and after registration using the motion fields estimated by NIFTI and UFCN are displayed in Fig. 4A. Bland-Altman analysis indicated UFCN achieved slightly higher NCC than NIFTI.Discussion
This study demonstrates the feasibility of using a novel unsupervised deep-learning method to estimate non-rigid respiratory motion fields from undersampled CMRA data. UFCN achieved comparable motion estimation to a widely used registration technique with 50 times faster computational speed. 2D non-rigid motion estimation for each slice of the 3D CMRA data is investigated first to reduce computational burden. Future studies will extend the network to enable 3D non-rigid respiratory motion estimation.Acknowledgements
No acknowledgement found.References
1. Prieto C, Doneva M, Usman M, Henningsson M, Greil G, Schaeffter T, Botnar RM. Highly Efficient Respiratory Motion Compensated Free-Breathing Coronary MRA Using Golden-Step Cartesian Acquisition. J Magn Reson Imaging 2015;41(3):738-746
2. Cruz G, Atkinson D, Henningsson M, Botnar RM, Prieto C. Highly efficient nonrigid motion-corrected 3D whole-heart coronary vessel wall imaging. Magn Reson Med 2017;77(5):1894-1908
3. Bustin A, Correia T, Rashid I, Cruz G, Neji R, Botnar RM, Prieto C. Highly Accelerated 3D Whole-Heart Isotropic Sub-Millimeter CMRA with Non-Rigid Motion Correction. 2019 ISMRM; Montreal, Canada. p0978
4. Henningsson M, Koken P, Stehning C, Razavi R, Prieto C, Botnar RM. Whole-heart coronary MR angiography with 2D self-navigated image reconstruction. Magn Reson Med 2012;67(2):437-445
5. Ilg E, Mayer N, Saikia T, Keuper M, Dosovitskiy A, Brox T. FlowNet 2.0: Evolution of Optical Flow Estimation with Deep Networks. 30th Ieee Conference on Computer Vision and Pattern Recognition (Cvpr 2017) 2017:1647-1655.
6. Meister S, Hur J, S. R. UnFlow: Unsupervised learning of optical flow with a bidirectional census loss. AAAI. New Orleans, Louisiana2018.
7. Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Transactions on Medical Imaging 1999;18(8):712-721.
Figures