3298
Use of population-derived perfusion information in predicting subject-specific perfusion references based on tissue partial volumes1Institute of Biomedical Engineering, Department of Engineering Science, University of Oxford, Oxford, United Kingdom, 2Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 3Department of Psychiatry, University of Oxford, Oxford, United Kingdom
Synopsis
Within and between subject variability in ASL perfusion data can obscure subtle perfusion changes indicative of neurodegenerative disease. It prohibits the production of a 'healthy' population-average perfusion atlas to which individuals can be compared. This work used a GLM to produce subject-specific perfusion references based on tissue partial volumes. Results show that such a linear model can successfully decompose perfusion, at the population level, into components related to tissue partial volumes. However, the model struggles to capture variability at the individual subject level and thus, the problem will in future be addressed with non-linear modelling techniques.
Introduction
Changes in cerebral tissue perfusion are associated with pathology [1,2]. Low SNR in ASL data and high inter-subject variability can obscure subtle perfusion changes resulting from neurodegenerative disease e.g. dementia [3,4]. This work considers whether structural imaging could be used to predict healthy perfusion in an individual, thereby generating personalised references against which measured perfusion could be compared. It is hypothesized that a personalised reference will be able to capture perfusion differences resulting from individual variations in cerebral anatomy.Methods
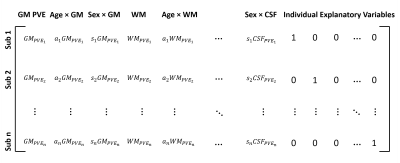
ASL data from 136 subjects included in the Whitehall II imaging sub-study [5] were used (19 female: 117 male, age range 62-81). The subjects were scanned using a multi PLD pcASL protocol (labelling duration=1.40s, PLDs: 0.25, 0.50, 0.75, 1.00, 1.25, 1.50, and 1.75s, TR=4.24s, TE=13ms, 24 slices each 4.95 mm thick, in-plane resolution 3.4mm x 3.4 mm, 112 volumes were acquired with 8 repeats at each PLD). Also acquired were a T1 structural image, head and body coil calibration images, and B0 fieldmaps. 116 individuals were included in a ‘training set’ and 20 were included in the ‘test set’. For each subject, perfusion analysis was performed using the FSL BASIL toolbox [6], which carries out motion correction, label-control subtraction, distortion and bias correction, kinetic model fitting, and calibration to produce perfusion images in absolute units (ml/100 g/min). Perfusion results were registered linearly to the native T1 space and non-linearly to MNI152 standard space, along with grey (GM) and white matter (WM) and CSF partial volume estimates generated with FSL’s FAST. Standard space calibrated perfusion maps for the n=116 training set subjects were concatenated and input to a General Linear Model (GLM) analysis in FEAT. A design matrix was constructed as shown in Figure 1, with GM, WM, and CSF partial volume estimates (PVEs) included as voxelwise explanatory variables (EVs), along with additional voxelwise EVs of demeaned age, $$$a_{i}$$$ , and demeaned sex, $$$s_{i}$$$ , each multiplied with each PVE map. An additional EV per subject was included in the design to encapsulate perfusion variation that could not be related to the voxelwise EVs through the GLM. After GLM fitting the model parameter estimates ($$$\beta$$$ values) associated with each EV were used to produce predicted perfusion maps in native ASL space for the n=20 test set individuals, according to the equation:$$CBF_{i}=CBF_{predicted_{i}}+CBF_{res_{i}}$$
$$CBF_{i}=GM_{PVE_{i}} \left(\beta_{GM}+a_{i}\beta_{GMa}+s_{i}\beta_{GMs}\right)+ WM_{PVE_{i}}\left(\beta_{WM}+a_{i}\beta_{WMa}+s_{i}\beta_{WMs}\right)\\+CSF_{PVE_{i}}\left(\beta_{CSF}+a_{i}\beta_{CSFa}+s_{i}\beta_{CSFs}\right)+ CBF_{res_{i}}$$
For each subject, $$$i$$$, a residual perfusion map, $$$CBF_{res_{i}}$$$ , was also generated by subtracting the predicted perfusion from the calibrated perfusion map, $$$CBF_{i}$$$, produced using BASIL. Pure grey matter (PVE>80%) and white matter (PVE>90%) ROIs were generated for each subject, and within-subject root-mean-squared RMS prediction errors were calculated according to the following equation:
$$GM_{error_{i}} = \left(\frac{1}{V_{i}}\sum_{v=1}^{v=V}\left[GM_{ROI}\cdot\left(CBF_{i}-CBF_{predicted_{i}}\right)\right]^{2}\right)^{\frac{1}{2}}$$
Where $$$V_{i}$$$ is the number of voxels within subject $$$i$$$ ’s brain mask.
Results
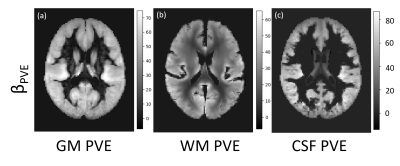
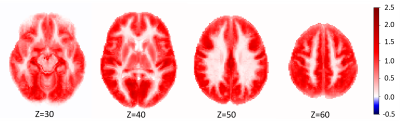
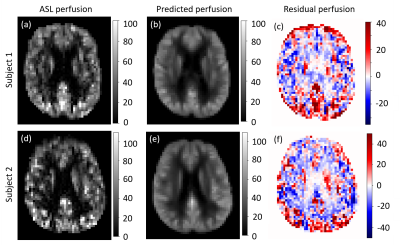
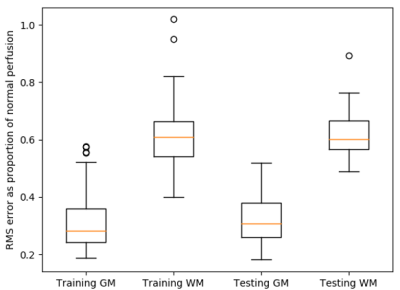
Figure 2 shows the parameter estimates ($$$\beta$$$ maps) associated with the 3 PVE map EVs, derived from the GLM. Figure 3 shows the mean of the 116 subject specific EVs, i.e. the mean $$$CBF_{res_{i}}$$$ for the training group. Voxelwise this represents perfusion that was not associated with age, sex, or any of the structural components. Averaged across the whole training set population (116 individuals), the magnitude of this residual was very low (<2.5 ml/100g/min as shown in Figure 3). However, for individuals within the training set the residuals could vary in the range $$$\pm50$$$ ml/100g/min. Figure 4 shows examples of perfusion estimated from ASL (Figure 4a,d) and perfusion predicted using the GLM model (Figure 4b,e), for two individuals from the test set. Figure 4 also shows the difference between the two perfusion images, demonstrating the voxelwise variability in perfusion residuals. Figure 5 shows prediction errors in grey matter and white matter voxels, as a proportion of ‘typical’ perfusion for each tissue (60 and 20 ml/100g/min for GM and WM, respectively) across both the training and test data sets. The figure shows similar performance in the training and test groups, but lower prediction error in ‘pure’ grey matter voxels compared with ‘pure’ white matter voxels.Discussion
This work developed a method for generating individual perfusion references, which accounts for voxelwise variability in cerebral anatomy. The images in Figure 2 show linear relationships between perfusion, and partial volume maps of GM, WM and CSF. It is possible that the apparent relationship between perfusion and CSF, a non-perfused tissue type, results from imperfect non-linear registration of training set perfusion and PVE images into standard space. The results in Figure 3 demonstrate that, across the training data set of 116 individuals, the GLM design in Figure 1 was able to model perfusion with very little average error. However, Figures 4 and 5 show that there is substantial variability in prediction error both voxelwise (Figure 4) and between individuals (Figure 5). It is likely that these differences result from a combination of factors, including cognitive performance, functional neural activation, and non-PVE structural features e.g. membership of a particular subcortical structure or degree of cortical folding.Conclusion
While the GLM successfully modelled perfusion at the population level, it was unable to represent perfusion at the individual subject level. Non-linear modelling techniques will be employed in future to better capture sources of variability in perfusion data.Acknowledgements
The Wellcome Centre for Integrative Neuroimaging is funded by a Centre Grant (203139/Z/16/Z). The authors gratefully acknowledge support from EPSRC grant: EP/P012361/1. The Whitehall II imaging sub-study was funded by UK Medical Research Council grant: G1001354. SS was supported by a fellowship award from Alzheimer's Society, UK (Grant Number 441).References
[1] Barker, P., Golay, X., and Zaharchuk, G., eds. Clinical perfusion MRI: techniques and applications. Cambridge University Press, 2013.
[2] Zhang, N., et al., "Cerebral blood flow measured by arterial spin labeling MRI at resting state in normal aging and Alzheimer’s disease.", Neurosci. & Biobehav. Rev. 2017 Jan;72:168-175.
[3] Chen, J., et al., "Age-associated reductions in cerebral blood flow are independent from regional atrophy." Neuroimage 2011 Mar 15;55(2):468-478.
[4] Petersen, E., et al., "The QUASAR reproducibility study, Part II: Results from a multi-center Arterial Spin Labeling test–retest study." Neuroimage 2010 Jan 1;49(1):104-113.
[5] Filippini, N., et al., "Study protocol: the Whitehall II imaging sub-study." BMC Psychiatry 2014 May 30;14:159.
[6] https://www.fmrib.ox.ac.uk/fsl/BASIL
Figures