3178
Local Structure Orientation (LSO) for histology-based validation of MRI: evaluation in a physical phantom study1Medical Imaging Sciences, Faculty of Health Sciences, University of Sydney, Sydney, Australia, 2Medical imaging sciences, King Saud University, Riyadh, Saudi Arabia, 3Biomedical Engineering, School of Aerospace Mechanical & Mechatronic Engineering, Faculty of Engineering and IT, University of Sydney, Sydney, Australia
Synopsis
Histology-based validation of magnetic resonance imaging (MRI) is essential to confirm imaging technique specificity and accuracy. One-to-one correspondence is complicated by histological preparation and differences in MRI and histology contrast mechanisms. A new approach, based on local structure orientation (LSO) and using microstructural features, is proposed. LSO facilitates registration based on direct correspondence between tissue fibre structure to emulate a common contrast mechanism. Using a physical phantom derived from a chicken heart with embedded fibrous structures, residual displacement was 14.1 mm for advanced mean squares (AMS), 2.2 mm for modified AMS (AMSR), and 3.3 mm for mutual information (MI).
Introduction
Precise registration of histology and MRI is confounded by fundamentally different contrast mechanisms, with image features visible in MRI and stained sections lacking one-to-one correspondence. Conventional solutions to this problem use landmarks and/or mutual information (MI) techniques (1-4). Landmark methods are limited by changed appearance or saliency of features in corresponding images. MI methods assume that pixel intensity distributions follow consistent relationships, and that corresponding texture features exist in both images, which may not be the case in some tissues (e.g. prostate). As a solution, we propose a new approach based on local structure orientation (LSO). LSO-registration uses tissue microstructure features (fibre orientation) that are measurable by both histology and MRI. These features provide a dense field for objective coregistration. In-plane fibre orientation angle can be generated from a histological image using a structure tensor analysis based on a method developed for neural fibre mapping (5), and from 3D or 2D diffusion tensor MRI (DTI). We validate the LSO-registration using a physical tissue structure phantom that simulates fibre orientation in a human prostate, the distinct MRI features (e.g. peripheral zone/transition zone boundary) of which show negligible contrast in stained histology sections.Methods
A. Simulation of human prostateWe used a chicken heart for its comparable size to a prostate and robust wall. The heart was filled with lightly cooked small pieces of chicken immersed in gelatin to produce a dense fiber field similar to that of prostate tissue, interspersed with a non-fibrous matrix.
B. MRI scans
The phantom was scanned using high resolution T2 weighted (T2w) and 2DTI sequences. LSO works in two dimensions, hence 2D DTI was used instead of 3D DTI to avoid signal noise from the additional dimension.
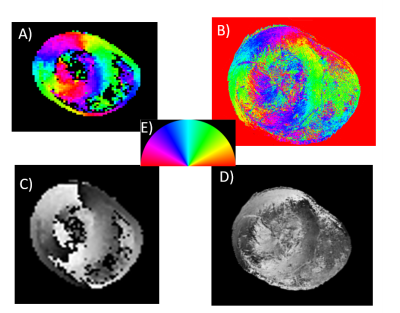
C. Generating a LSO map from histology
The phantom was processed using conventional histological techniques (fixation, sectioning, and digitization). Using a mould-based system for consistent and reproducible sectioning(6), we selected a matching histological slice and image slice. A LSO map of the slice was generated using a structure tensor algorithm based on a method developed for neural fibre mapping (5). The LSO map was down-sampled to the same size as the T2w (256x256).
D. Registration
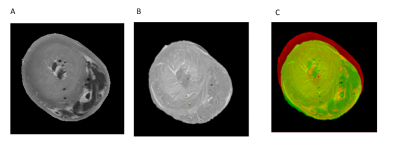
Elastix (7) was used to register the DTI map (fixed image) and the histology LSO map (floating image) (Figure 1). Registration involved an affine transformation followed by a B-spline transformation to account for non-rigid deformation arising from histological techniques. Three cost functions were tested: advance mean squares (AMS), and a modified advanced mean squares (AMSR) to correct angular redundancies and MI. Angular redundancies were corrected using the AMSR function by converting orientation angle differences >p/2 to the complement (p - angle). The registration output was the aligned LSO image and the estimated registration transformation matrices. The third cost function, MI, was tested by registering T2w to the corresponding histology image (Figure 2).
E. Assessment of registration accuracy
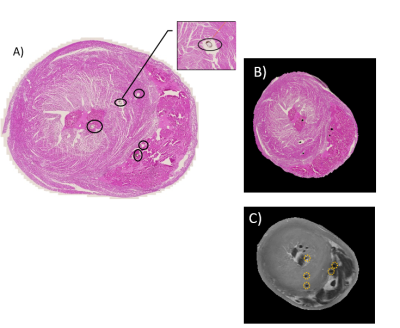
Fiducial markers were designed that were visible on both the T2w and corresponding histology slice images. We inserted 8 thin cotton threads covered with gold acrylic paint using a size 15 beading needle. Five markers were visible on the T2w and aligned histology image (Figure 3). We measured the mean displacement in millimetres between the original marker locations on the T2w and the aligned locations on the registered histology section.
Results and Discussion
The LSO-based registration method using the modified cost (AMSR) outperformed other approaches, demonstrating the importance of correcting for fibre orientation redundancy. Before registration, the fiducial marker displacement was 12.0 mm. After registration, residual displacement was 2.2 mm for AMSR, 14.1 mm for AMS, and 3.3 mm for MI.Interestingly, visual assessment of the alignment between T2w and histology indicated that MI had better apparent global alignment than AMSR. One limitation of the current approach is that the threaded fiducials are not localised throughout the sample. This method will be refined to ensure that the displacements we are measuring are truly representative of the global registration accuracy.
LSO facilitates registration based on direct correspondence between image features acquired from histology and MRI, and works even at low DTI resolution. Both the concept of co-registering MRI and histology based on a shared contrast mechanism, and the physical phantom used in this work, are entirely new, and when combined they represent an innovative method for validating MRI using histology. In future work we will investigate the robustness of the method to different deformations.
Conclusion
We describe a new method to co-register MRI and histology data which exploits tissue fibre structure to emulate a common contrast mechanism. The method was tested using a physical phantom simulating the prostate and outperformed conventional registration based on MI. Our preliminary investigations of LSO are being extended to include real human prostate. We consider the potential for applying the LSO method in clinical studies with fibrous tissues, such as the heart or prostate, to be promising.Acknowledgements
No acknowledgement found.References
1. Goubran M, de Ribaupierre S, Hammond RR, Currie C, Burneo JG, Parrent AG, et al. Registration of in-vivo to ex-vivo MRI of surgically resected specimens: A pipeline for histology to in-vivo registration. Journal of Neuroscience Methods. 2015;241:53-65.
2. Gibson E, Crukley C, Gaed M, Gomez JA, Moussa M, Chin JL, et al. Registration of prostate histology images to ex vivo MR images via strand-shaped fiducials. Journal of magnetic resonance imaging : JMRI. 2012;36(6):1402-12.
3. Orczyk C, Mikheev A, Rosenkrantz AB, Melamed J, Taneja SS, Rusinek H. Imaging of prostate cancer: a platform for 3D co-registration of in-vivo MRI ex-vivo MRI and pathology. Proceedings of SPIE. 2012;8316:83162M.
4. Antunes J, Viswanath S, Brady JT, Crawshaw B, Ros P, Steele S, et al. Coregistration of Preoperative MRI with Ex Vivo Mesorectal Pathology Specimens to Spatially Map Post-treatment Changes in Rectal Cancer Onto In Vivo Imaging: Preliminary Findings. Academic radiology. 2018;25(7):833-41.
5. Grussu F, Schneider T, Yates RL, Zhang H, Wheeler-Kingshott C, DeLuca GC, et al. A framework for optimal whole-sample histological quantification of neurite orientation dispersion in the human spinal cord. Journal of neuroscience methods. 2016;273:20-32.
6. Bourne RM, Bailey C, Johnston EW, Pye H, Heavey S, Whitaker H, et al. Apparatus for Histological Validation of In Vivo and Ex Vivo Magnetic Resonance Imaging of the Human Prostate. Frontiers in Oncology. 2017;7(47).
7. Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging. 2010;29(1):196-205.
Figures