3030
Comparison of Selective Excitation and IDEAL for Chemical Shift Imaging of Hyperpolarized [1-13C]Pyruvate
Keith A Michel1, Christopher M Walker1, Matthew E Merritt2, and James A Bankson1
1Imaging Physics, The University of Texas MD Anderson Cancer Center, Houston, TX, United States, 2Biochemistry and Molecular Biology, University of Florida, Gainesville, FL, United States
1Imaging Physics, The University of Texas MD Anderson Cancer Center, Houston, TX, United States, 2Biochemistry and Molecular Biology, University of Florida, Gainesville, FL, United States
Synopsis
Imaging of hyperpolarized (HP) agents permits real-time non-invasive measurement of metabolic flux, however these experiments require specialized techniques for rapidly encoding chemical shift in a way that is sensitive metabolic signals that are often low SNR. The SNR performance of two chemical shift encoding methods commonly used for HP 13C MRI, IDEAL and spectral-spatial excitation pulses, were compared for a typical preclinical experiment imaging [1-13C]pyruvate at 7 T. The SNR of these two techniques were found to be equivalent in numerical simulation, while the SNR measured for IDEAL was ~30% greater than for spectral-spatial pulses in a thermal phantom experiment.
Introduction
Dissolution dynamic nuclear polarization of 13C agents enables rapid measurement of cellular metabolism in vivo.1 These measurements are challenging due in part to the transient nature of hyperpolarized (HP) signals, which persist for only around a minute in tissue and vary continuously due to the effects of perfusion, cellular uptake and metabolism. A typical HP experiment must therefore dynamically encode chemical shift information using specialized imaging techniques.A variety of chemical shift encoding strategies have been used for imaging of metabolic agents by HP 13C MRI. Among the most widely used spectroscopic encoding methods for this purpose are spectrally selective RF pulses and multi-echo chemical shift decomposition (IDEAL). Specialized excitations, for example using compound spectral-spatial pulses, can perturb a single metabolite for selective imaging.2 This approach is simple in practice, however the design of these pulses can be challenging depending on the spectral response needed and the capabilities of a given MRI system. Alternately, traditional broadband excitations can be used with spectroscopic readouts to obtain individual chemical shift images. Among spectroscopic readout strategies, IDEAL is appealing since it constrains encoding in a way that is optimized for the specific set of chemical shifts that are known to be present in a given HP experiment.3 In this work, we compare the SNR performance of selective excitations and IDEAL readouts for a typical preclinical imaging experiment using [1-13C]pyruvate.
Methods
We compared SNR performance of IDEAL and a spectral-spatial pulse designed for selective imaging of [1-13C]pyruvate and [1-13C]lactate at 7 T in numerical simulation and phantom experiments. In phantom experiments, IDEAL and spectral-spatial data were acquired for a range of effective excitation angles from a [13C]urea phantom using custom-built methods on a Bruker 7T MRI system equipped with BGA12-SHP gradients capable of delivering up to 660 mT/m and 4500 T/m/s. Numerical simulations were conducted in MATLAB to simulate data acquired with identical timing as phantom data, but with a range of effective excitation angles, T2* relaxation time constants and variances of complex zero-mean Gaussian noise in k-space data.The 6.6 ms flyback spectral-spatial pulse considered in this work (Figure 1) was designed for individual excitation of pyruvate or lactate using a publicly available MATLAB toolbox.2 An IDEAL encoding scheme using a series of eight 1.6 ms, 5 kHz SLR pulses with a TE shift of 0.52 ms was chosen for optimal signal averaging in the resulting chemical shift images (Figure 2). The same 2D singleshot flyback EPI imaging readout was used for both chemical shift encoding schemes (Figure 3). IDEAL was applied to raw k-space data with correction for chemical phase evolution along each imaging readout.4 No fieldmap demodulation was applied, and without this correction IDEAL is a linear transformation from the temporal to spectral domain with a magnitude and phase response shown in Figure 2. IDEAL data were acquired with minimum inter-echo delays to minimize T1 effects.
In order to facilitate comparison to individual spectral-spatial pulses, a variable excitation angle scheme was used for IDEAL across the eight pulses applied in order to provide a consistent level of transverse magnetization (MZ) without consideration of relaxation effects. Individual excitation angles (θ) were calculated from the desired effective excitation angle (Θ) imparted by the series of N pulses as:5
$$\theta_i = \arcsin \left( \frac{\sin \Theta}{\sqrt{N} M_{Z,i-1}} \right) = \arcsin \left( \frac{\sin \Theta}{\sqrt{N} \prod^{i-1}_{j=0} \cos \theta_j} \right)$$
Results
The SNR performance of our single-band spectral-spatial RF pulse and our eight echo IDEAL encoding strategy are summarized in Figure 4. The SNR of images obtained by selective excitation and IDEAL encoding were found to be approximately identical in numerical simulations. Under no conditions were the mean SNR values of selective excitation and IDEAL images separated by greater than one standard deviation about their respective means. In thermal phantom experiments, IDEAL encoding consistently demonstrated a greater SNR than spectral-spatial excitation. Across all effective excitation angles measured, the mean ratio in mean SNR obtained by IDEAL relative to selective excitation was 32%. The greatest ratio in mean SNR measured for IDEAL relative to selective excitation was 42% at an effective excitation angle of 20 degrees.Discussion
MRI scan parameters for HP experiments must be set carefully to provide optimal performance given the non-renewable nature of the imaging signal and the relatively low amplitude of signals produced via metabolic conversion in vivo. In this work we compared the SNR performance of two commonly used chemical shift encoding strategies for preclinical HP MRI experiments using [1-13C]pyruvate. A vital consideration in comparing these spectroscopic imaging strategies was the concept of effective flip angle, which captures the amount of longitudinal magnetization expunged to generate a set of images. While very similar SNR values were seen in numerical simulation, IDEAL demonstrated an SNR improvement of about 32% in thermal phantom experiments. Further study is needed to determine the reason for the greater SNR of IDEAL imaging in phantom experiments. Lastly, we note that the results presented are subject to the capabilities of the preclinical 7 T MRI system used in this work. Different results may be expected for MRI systems operating at different field strengths or with different gradient capabilities.Acknowledgements
This work was supported in part by the National Institues of Health (R01-DK105346, R01-CA211150, P30-CA016672). The content represents the views of the authors and not of any sponsoring organization.References
- Kurhanewicz J, Vigneron DB, Brindle K, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia 2011;13(2):81-97.
- Larson PE, Kerr AB, Chen AP, et al. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson 2008;194(1):121-127.
- Reeder SB, Brittain JH, Grist TM, Yen YF. Least-squares chemical shift separation for (13)C metabolic imaging. J Magn Reson Imaging 2007;26(4):1145-1152.
- Brodsky EK, Holmes JH, Yu H, Reeder SB. Generalized k-space decomposition with chemical shift correction for non-Cartesian water-fat imaging. Magn Reson Med 2008;59(5):1151-64.
- Nagashima K. Optimum pulse flip angles for multi-scan acquisition of hyperpolarized NMR and MRI. J Magn Reson 2008;190(2):183-188.
Figures
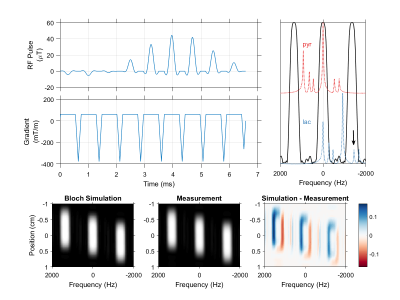
The spectral-spatial pulse used in this work, designed using a publicly available MATLAB package.2 The excitation waveform has a total duration of 6.6 ms and slice thickness of 1 cm. A 420 Hz passband and 930 Hz stopband allow selective imaging of pyruvate and lactate at 7 T (top right). Urea at -1680 Hz (arrow) falls within a passband alias when the MRI center frequency is set to lactate, but can be pre-saturated when using this pulse for dynamic imaging. Bloch simulation of the transverse magnetization response to this pulse agrees with measurement in a relaxed water phantom (bottom row).
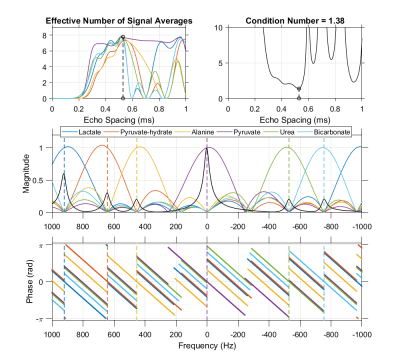
An echo spacing of 0.52 ms with 8 echoes was chosen for IDEAL encoding of [13C]urea, [1-13C]pyruvate and associated metabolites at 7 T. This echo spacing minimizes the condition number of the IDEAL phase coefficient matrix and optimizes noise performance in the resulting individual chemical shift images. Without fieldmap demodulation, this IDEAL decomposition is a linear transform whose frequency response is characterized in the magnitude and phase plots shown here. Dashed vertical lines indicate the center frequency of each resonance at 7 T.
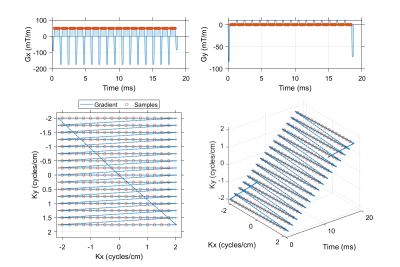
A flyback EPI readout with a 16 x 16 matrix, 4 cm field of view and 20.833 kHz readout bandwidth (48 µs dwell time) was used in phantom and simulation experiments of spectral encoding methods.
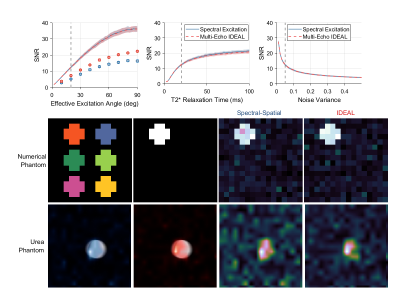
Results of simulation and phantom experiments comparing spectral encoding strategies for imaging of [1-13C]pyruvate and its associated metabolites at 7 T. The mean output SNRs for a single metabolite is summarized as a function of effective excitation angle, T2* relaxation time constant and noise variance in acquired complex data (top row). Very similar noise performance is achieved for HP imaging in simulations with spectrally selective excitations and IDEAL encoding, however the mean SNR measured in a 13C urea phantom was consistently ~30% greater for IDEAL encoding (circles).