2966
Accelerating volumetric 31P MRSI of the human calf muscle at 7 Tesla: can low-rank denoising filters replace the need for signal averaging?1Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany
Synopsis
Volumetric 31P MRSI in the human calf muscle at 7T in principle enables acquisitions with high spatial resolution, but may require signal averaging to improve signal-to-noise ratios of low-concentrated metabolites. To minimize the acquisition duration of volumetric 31P MRSI, in this study we demonstrate that the acquisition of signal averages can be reliably substituted by application of low-rank denoising filters without the introduction of a quantification bias. This allows acquisition of 31P MRSI data with 1 ml voxels in measurement durations of only 14 minutes that have the same quality as if acquired in 56 minutes with signal averaging.
Introduction
In principle, the sensitivity of volumetric 31P MRSI in the human calf muscle at 7T1 makes it feasible to acquire small voxel volumes in the range of 1 ml. Unfortunately, the signal-to-noise ratio (SNR) of low-concentrated metabolites like inorganic phosphate (Pi) is still often poor at high spatial resolutions, requiring the acquisition of signal averages. However, even a small number of averages translates to long measurement times, e.g. on the order of one hour for 3D chemical shift imaging (CSI) with large matrix sizes (> 16³ voxels).An alternative to signal averaging for SNR enhancement could be the utilization of the low-rank properties of MRSI datasets by means of denoising filters2. Though having the potential to accelerate volumetric MRSI acquisitions, low-rank filtering bears the risk of biasing quantified spectral parameters if performed excessively.
The purpose of this study is to verify that the acquisition of signal averages in volumetric 31P MRSI of the human calf muscle at 7T can reliably be substituted with the application of low-rank denoising filters to reduce the total measurement time.
Methods
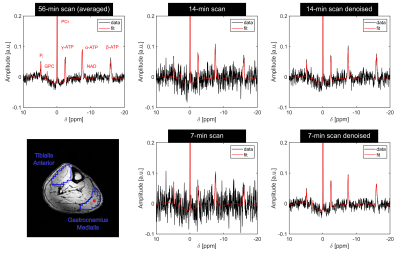
Four healthy volunteers (3 male / 1 female, age: 25-32) were examined on a 7-T whole-body MR system (Siemens Healthineers) with a double-resonant 31P-1H volume resonator (RAPID Biomedical). 31P MRSI datasets of the calf muscles were acquired using an acquisition-weighted 3D CSI sequence (matrix size $$$=~24\times24\times16$$$, spatial resolution $$$=~(8\times8\times16)~\textrm{mm}^3$$$, $$$T_R~=~240~\textrm{ms}$$$, $$$\alpha~=~20°$$$, $$$\Delta{f}~=~5000~\textrm{Hz}$$$, 1024 data points). The acquisition duration for the averaged, Hamming-weighted acquisition with 16 averages in the k-space center was 56 minutes (56-min scan).To obtain synthetic datasets of a faster measurement (14-min scan), only the first average of each acquired k-space point in the 56-min scan was reconstructed. Gaussian white noise was added to the 14-min scan to synthesize a 2-fold accelerated measurement (7-min scan). A k-space filter function corresponding to the averaging scheme was applied to both the 14- and 7-min scans to obtain the same effective spatial resolution as in the 56-min scan. Denoising of these synthetic MRSI datasets was performed via a low-rank approximation of the spatial-spectral Casorati matrix according to the singular value hard thresholding criterion of Gavish and Donoho3.
All obtained 31P MRSI datasets were processed by one-fold spatial zero-filling and application of a 10-Hz Gaussian filter in the time domain, and corrected for zero- and first-order phases. Localized 31P spectra were evaluated for Phosphocreatine (PCr), Adenosine-5’-Triphosphate (ATP), Nicotinamide-Adenine-Dinucleotide (NAD), Pi, and Glycerophosphocholine (GPC) using a customized Matlab (The Mathworks) implementation of the AMARES algorithm4. pH values were calculated employing the modified Henderson-Hasselbalch equation5 to the chemical shift difference between PCr and Pi.
Results
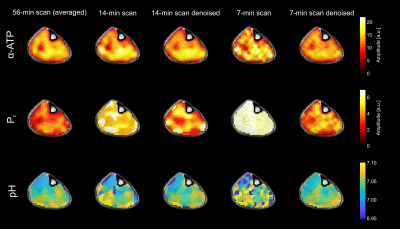
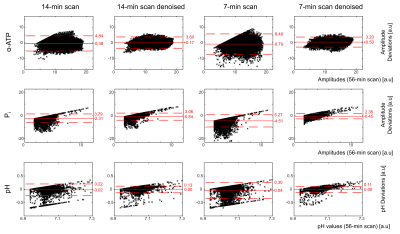
In all volunteers, localized 31P spectra obtained from the 56-min scan had sufficient SNR for a robust quantification of low-concentration metabolites, i.e. Pi (Figure 1). With denoising, SNR increased by a factor of 1.68±0.01 and 3.71±0.07 (thresholding ranks 109±1 and 29±1; mean across volunteers) for the 14- and 7-min scans, respectively, enabling the robust quantification of the Pi resonance also in the synthetic datasets of all volunteers.The metabolite and pH maps visually demonstrate that the quantification results of the 14- and 7-min scans with denoising resemble the results obtained with the 56-min scan (Figure 2). Quantitatively, the results of the 14- and 7-min scans with denoising showed consistently reduced bias and deviation from the results of the 56-min scan, compared to the results without denoising (Figure 3).
The mean pH values across the anterior tibialis and medial gastrocnemius muscles obtained from all datasets of all volunteers show that also the local pH differences observable in the 56-min scans are preserved in the 14- and 7-min scans with denoising (Table 1).
Discussion
All quantification results of the denoised datasets agree with the averaged datasets within the quantification errors (Cramer-Rao Lower Bounds; not shown); thus, the applied acceleration strategy proves to be valid. This strategy improves quantification of higher SNR signals, such as e.g. α-ATP, and enables a more reliable quantification of low SNR signals (e.g. Pi amplitude no longer runs into the fitting boundaries).The denoising capabilities with negligible quantification bias in this study can be attributed to (I) the high SNR of volumetric 31P MRSI in the human calf muscle at 7T, and (II) the large number of voxels (matrix size > 16³) available for efficient filtering. It should be noted that low-rank filtering of acquisitions with lower SNR could introduce higher quantification bias. The limits of the acceleration strategy with respect to (I) the lowest required SNR of the MRSI acquisition and (II) possible SNR gains with negligible quantification bias have to be investigated in future simulation studies.
Additionally, the results obtained from the denoised 7-min scan suggest that the proposed strategy remains valid even for noisier 31P MRSI datasets, enabling potentially further acceleration using fast MRSI sequences like echo-planar spectroscopic imaging1.
Conclusion
Low-rank denoising filters can reliably replace signal averaging to reduce the acquisition duration of volumetric 31P MRSI of human calf muscles at 7T. For the experimental setup utilized here, this concept proved to speed up acquisitions about a factor of 4 and enabled the acquisition of high-quality 3D 31P MRSI datasets with voxel sizes of about 1 ml in 14 minutes.Acknowledgements
No acknowledgement found.References
1 Korzowski A, Bachert P. High‐resolution 31P echo‐planar spectroscopic imaging in vivo at 7T. Magn Reson Med 2018;79:1251–1259.
2 Liang ZP. Spatiotemporal imaging with partially separable functions. In IEEE International Symposium on Biomedical Imaging, Arlington, VA, USA, 2007. pp. 988–991.
3 Gavish M, Donoho DL. The Optimal Hard Threshold for Singular Values is (4/√3). IEEE Trans. Inf. Theory 2014;60:5040–5053.
4 Vanhamme L, et al. Improved Method for Accurate and Efficient Quantification of MRS Data with Use of Prior Knowledge. J Magn Reson 1997;129:35–43.
5 De Graaf RA. In vivo NMR spectroscopy, principles and techniques, 2nd ed. Chichester, United Kingdom: Wiley, 2007.
Figures