2934
Metabolomic profiling of wildtype and transgenic Giardia lamblia strains by 1H Magic Angle Spinning NMR spectroscopy.
Martina Vermathen1, Norbert Mueller2, David Leitsch3, Damian Hertig4, Peter Vermathen4, and Joachim Mueller2
1Department of Chemistry and Biochemistry, University Bern, Bern, Switzerland, 2Institute of Parasitology, Vetsuisse Faculty, University Bern, Bern, Switzerland, 3Institute of Specific Prophylaxis and Tropical Medicine, Medical University of Vienna, Vienna, Austria, 4DBMR & DIPR, University Bern, Bern, Switzerland
1Department of Chemistry and Biochemistry, University Bern, Bern, Switzerland, 2Institute of Parasitology, Vetsuisse Faculty, University Bern, Bern, Switzerland, 3Institute of Specific Prophylaxis and Tropical Medicine, Medical University of Vienna, Vienna, Austria, 4DBMR & DIPR, University Bern, Bern, Switzerland
Synopsis
The intestinal protozoan parasite Giardia lamblia is a major cause of persistent diarrhea in humans and animals worldwide. Treatment of giardiasis with nitro-prodrugs relies on intrinsic enzymes in G.lamblia that are involved in activating or deactivating the prodrugs. To elucidate more about the physiologic function of these pathways and their potential roles in drug resistance, G.lamblia wildtype and transgenic strains were metabolomically characterized based on 1H HR-MAS NMR data and specific metabolites that may be related to nitroreductase activity were identified and discussed.
INTRODUCTION
The intestinal anaerobic protozoan parasite Giardia-lamblia is a major cause of persistent diarrhea. The treatment of Giardia infections is typically performed with nitro-compounds1. Most likely the mechanism of action relies on the reduction of the nitro group via enzymatic pathways in G.lamblia thereby activating the prodrugs. Enzymes such as thioredoxin reductase (TrxR) and the nitroreductases NR1, NR2, and NR3 with weak NR activity have been identified in G.lamblia and may be involved in the activation of or the resistance towards nitro-prodrugs2,3. Overexpression of NR1 results in an increased susceptibility to nitro-compounds in G.lamblia, whereas overexpression of NR2 results in an increased resistance in E.coli2,3. TrxR was found to have only little impact on nitro-drug activity and TrxR overexpression in resistant strains may be related to its role as radical scavenger4. The physiologic functions of these enzymes TrxR and NR1-3 in G.lamblia are not yet known.Previously, we have introduced HR-MAS NMR to study the metabolome of G.lamblia5. The aim of the current study was to compare the metabolomic patterns of G.lamblia strains with altered expression levels of the respective genes for getting insight into physiologic pathways.
EXPERIMENTAL PROCEDURES
G.lamblia strains -(i) G.lamblia overexpressing either the wildtype thioredoxin reductase (TrxR, n=10), or a dominant negative allele (TrxR-DN, n=10) and WBC6 wildtype as controls (WT1, n=10);
(ii) G.lamblia overexpressing the nitroreductases NR1 (n=5), NR2 (n=6), and NR3 (n=5), control strains overexpressing glucuronidase A (GusA, n=6) and WBC6 wildtype (WT2, n=4);
(iii) G.lamblia nitro-drug resistant strain (C4, n=10) and WBC6 wildtype (WT3, n=4).
Preparation of cell samples - G.lamblia trophozoites were grown to confluence in modified TYI-S33 medium and harvested. After centrifugation, washing, and shock freezing, samples were stored at -70°C until measurement.
NMR Spectroscopy - NMR was performed on a 500 MHz NMR spectrometer. 1H HR-MAS NMR experiments (3kHz and 279K) were recorded using a 1D-PROJECT sequence6 with water-presaturation.
Data Analysis - The NMR spectra were subdivided into 138 individually sized buckets. Multivariate data analysis was performed applying probabilistic quotient normalization (PQN), mean centering and pareto scaling. PCA (for unsupervised detection of group clustering) and orthogonal partial least squares discriminant analysis (oPLS-DA) were performed, cross-validated, and subjected to permutation testing.
For univariate statistical analysis representative spectral regions for 27 metabolites were integrated, normalized and mean±sd calculated. Single factor ANOVA and t-tests were performed. p-values were multiplied with 27 to correct for multiple comparisons (Bonferroni-correction). Metabolites that were significantly different between the independent wildtype populations were excluded from the discussion (Ala, Asn, Glu, Leu, Thr, Val, Glc-1-P, and TMA).
RESULTS
Three independent sample cohorts of G.lamblia trophozoites were compared:(i) TrxR, TrxR-DN, and WT1;
(ii) NR1, NR2, NR3, GusA, and WT2;
(iii) C4 and WT3.
Twenty-seven metabolites were identified based on our previous assignment5. PCA and oPLS-DA score plots and average normalized peak integrals for each of the 27 metabolites are displayed in Fig.1 for the first and in Fig.2 for the second sample cohort. The two transgenic strains TrxR and TrxR-DN were well separated from WT1 in both, PCA and oPLS with TrxR scores lying furthest away from WT1 (Fig.1A,B). Main metabolite contributions to distinguishing TrxR and TrxR-DN derived from increased acetate and decreased lysine levels in TrxR (Fig.1C).
When comparing trophozoites overexpressing nitroreductases NR1, NR2, and NR3 to GusA and WT2, PCA and oPLS-DA resulted in complete separation of NR1 and NR2 classes whereas NR3, GusA and WT2 overlapped (Fig.2A,B). The plot shows greatest distance for NR1 from WT and there is the order NR1 -> NR2 -> NR3 clustering with GusA -> WT. Specifically, nine metabolites were significantly altered in NR1 versus GusA (Fig.2C), namely acetate, cystine, glutamine, methionine, phenylalanine, tyrosine, citrulline, ornithine and pipecolic-acid. A typical NMR spectrum representing peaks for ornithine and citrulline from NR1 and GusA trophozoites is shown in Fig.3. To probe, whether the metabolic pattern obtained by comparing NR1 versus GusA could be linked to the nitro drug susceptibility, an oPLS-DA model calculated for discrimination between NR1 and GusA (Fig.4A) was applied to the nitro drug resistant C4 trophozoites with reduced NR1 activity and WT (Fig.4B). The predictive value of the model was very good, leading to complete separation of C4 versus WT, i.e. suggesting a correlation between nitroreductase activity and the metabolome. This was underlined by a significant correlation between the ratios WT/C4 vs. NR1/GusA of eight metabolites (Fig.4C).
DISCUSSION
Trophozoites expressing the nitroreductase NR1 exhibited a distinct pattern of nine differentially regulated metabolites. This pattern inversely correlated with a pattern of the same metabolites in the nitro drug resistant strain C4. The catabolism of arginine, a major energy source for G.lamblia, is of particular interest. Citrulline and ornithine, the first and the second catabolites of arginine were positively, glutamine, an indirect catabolite of ornithine, was negatively correlated with NR1 levels, suggesting that NR1 interferes with the catabolism of arginine. Since the ATP levels are unaltered in NR1 as well as in all other transgenic trophozoites, a direct interference with energy supply is unlikely.CONCLUSION
The study demonstrates significant metabolic differences between G.lamblia wildtype and transgenic strains that are likely related to nitroreductase activity.Acknowledgements
This work was supported by the Swiss National Science Foundation (grant No. 31003A_163230) and grant No. 206021-128736 for the purchase of the HR-MAS probe.References
- Leitsch, D. A review on metronidazole: an old warhorse in antimicrobial chemotherapy. Parasitology. 2019; 146:1167-1178.
- Müller, J.; Rout, S.; Leitsch, D.; Vaithilingam, J.; Hehl, A.; Müller, N. Comparative characterisation of two nitroreductases from Giardia lamblia as potential activators of nitro compounds. Int J Parasitol Drugs Drug Resist. 2015; 5:37-43.
- Müller, J.; Müller, N. Nitroreductases of bacterial origin in Giardia lamblia: Potential role in detoxification of xenobiotics. MicrobiologyOpen 2019, 10.1002/mbo3.904, e904, doi:10.1002/mbo3.904.
- Müller, J.; Braga, S.; Heller, M.; Müller, N. Resistance formation to nitro drugs in Giardia lamblia: No common markers identified by comparative proteomics Int J Parasitol: Drugs Drug Resist. 2019; 9:112-119.
- Vermathen, M.; Müller, J.; Furrer, J.; Müller, N.; Vermathen, P. 1H HR-MAS NMR spectroscopy to study the metabolome of the protozoan parasite Giardia lamblia. Talanta. 2018; 188:429-441.
- J.A. Aguilar, M. Nilsson; G. Bodenhausen G.A. Morris. Spin echo NMR spectra without J modulation. Chem. Commun. 1012; 48:811-813.
Figures
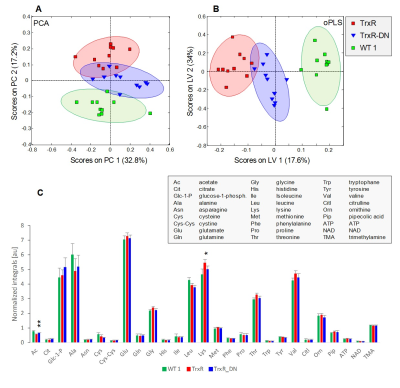
(A)
PCA and (B) oPLS-DA score plots for
TrxR, TrxR-DN and WT1. Ellipses correspond to 83.4% confidence intervals. (C) Bar plots displaying mean
normalized integrals ± SD derived from representative spectral regions for 27
metabolites in TrxR, TrxR-DN and WT1. Significance test was performed comparing
TrxR and TrxR-DN with * p<0.05 and ** p<0.01. Metabolites that exhibited
significant differences between independent WT cohorts were excluded from
testing.
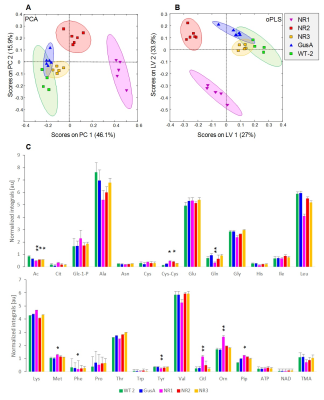
(A) PCA and (B) oPLS-DA score plots for NR1, NR2,
NR3, GusA and WT2. Ellipses correspond to 83.4% confidence intervals. (C) Bar plots displaying mean
normalized integrals ± SD derived from representative spectral regions for 27
metabolites in NR1, NR2, NR3, GusA, and WT2. Significance test was performed
comparing NR1-3 versus GusA with * p<0.05 and ** p<0.01. Metabolites that
exhibited significant differences between independent WT cohorts were excluded
from testing.
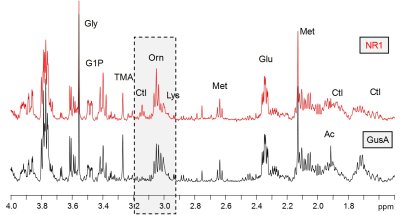
1H
HR-MAS NMR spectra (1.5 ppm – 4.0 ppm) of Giardia trophozoite suspensions in
PBS (D2O, pH 7, T = 6°C) obtained from GusA (black) and NR1 (red)
strains. The grey box highlights the ornithine and citrulline resonances that
had significantly different levels in NR1 and GusA.
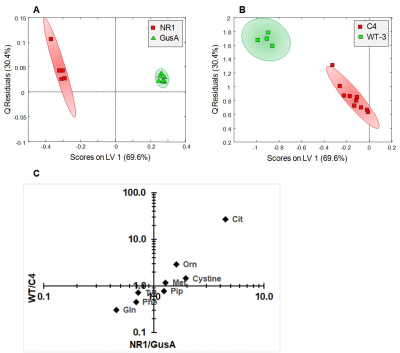
(A)
oPLS-DA score plots displaying the model
calculated for distinguishing NR1 from GusA along the latent variable LV1. (B) Class prediction based on the model
shown in (A) applied to C4 and WT3 strains. (C) Relative changes observed in NR1 compared to GusA of the eight
differential metabolites versus the corresponding relative changes obtained in
WT compared to C4. The correlation between metabolite ratios in NR1 vs. GusA
and WT vs. C4 trophozoites is highly significant (p<0.001).