2913
Rapid acquisition of semi-LASER MR spectroscopic imaging using symmetric EPSI encoding: A preliminary study in phantom and human brain1Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Brigham and Women’s Hospital, Boston, MA, United States, 3MR Applied Science Lab, GE Healthcare, Berlin, Germany
Synopsis
The aim of this study was to develop and test rapid MR spectroscopic imaging (MRSI) in patients with brain tumors. We implemented and optimized rapid MRSI at 3T using semi-localization by adiabatic selective refocusing pulses (semi-LASER), fast spatial encoding using symmetric echo planar spectroscopic imaging (EPSI), and robust water suppression with variable power and optimized relaxation delays. This rapid imaging approach was tested in a phantom and subsequently in a volunteer and a patient. In vivo studies demonstrated that high quality MRSI data with voxel volumes less than 0.5 mL can be collected within a clinically feasible time (2 minutes).
Introduction
Magnetic resonance spectroscopic imaging (MRSI) is an important technique for assessing the spatial variation of metabolites in vivo. In brain cancer imaging, PRESS and STEAM sequences are used routinely in single-voxel and multi-voxel spectroscopic imaging, with special interest in differentiating radiation necrosis from tumor recurrence and in monitoring treatment response. In PRESS, refocusing pulses have smaller bandwidths and are thus prone to introducing chemical shift displacement errors. These errors can be reduced with semi-localization using adiabatic pulses (semi-LASER or sLASER) (1), where refocusing pulses have broad bandwidths that enable precise volume localization with minimal or no chemical shift displacement errors. In addition, water can be suppressed using a VAPOR scheme (2).The clinical applicability of conventional MRSI has been limited due to long scan times which increase patient discomfort and increase motion artifacts. Hence, rapid acquisition strategies for MRSI are of great clinical interest (3). At an ultra-high field of 7T, symmetric echo planar spectroscopic imaging (EPSI) has recently shown to promising within a clinically feasibile scan time compared with flyback EPSI (4). The aim of this study was to implement and optimize MRSI at 3T using rapid EPSI spatial encoding in patients with brain tumors.
Methods
Experiments were performed in an MRS phantom (GE Braino MRS) as well as in-vivo using a 3T clinical scanner (MR750w, GE Healthcare) with an 8-channel head coil. The volunteer and patient in this study provided written informed consent under an IRB-approved prospective protocol.Anatomical T2-weighted FLAIR (TR/TE=9000/120ms; flip angle=90; 320x256; FOV=24; slice thickness=3mm; acquisition time:3-4min) and single-slice MRSI were performed using conventional phase encoding PRESS sequence with TR / TE = 2 s / 35 ms. Water suppressed sLASER-EPSI was performed using an 8x8 and 16×16 matrix under different experimental conditions as shown in Table 1. A radiologist assessed the image quality of MRSI. Also, water reference scan with full encoding was obtained for eddy current correction. Prior to the MRSI acquisition, power calibration of VAPOR was performed for each subject to improve water suppression. Water scans were acquired to correct the odd and even echoes of the symmetric EPSI sequence with MRSI data.
All raw MRSI data and images were transferred to an offline workstation. Data was filtered using 4 Hz exponential filtering, and an in-house MATLAB code was used to generate MRSI spatial and overlay maps.
| Table1: Sequence parameters used for phantom, volunteer, and patient scans | | | | | ||||||
| | | | | | | | | | | |
| Localiztion sequence | Encoding type * | Scan | TR (msec) | FOV, mm | Matrix size | Slice thickness, mm | Voxel size (mm3) | Volume (mL) | NEX | Scan time (min:sec) |
| PRESS | PE | Phantom | 2000 | 80 × 80 | 8 × 8 | 10 | 10 × 10 × 10 | 1 | 2 | 4:20 |
| Semi-LASER | Symmetric EPSI | Phantom | 2000 | 80 × 80 | 8 × 8 | 10 | 10 × 10 × 10 | 1 | 2 | 0:40 |
| Semi-LASER | Symmetric EPSI | Phantom | 2000 | 90 × 90 | 16 × 16 | 15 | 6 × 6 × 10 | 0.36 | 2 | 1:04 |
| Semi-LASER | Symmetric EPSI | Volunteer | 2000 | 90 × 90 | 16 × 16 | 10 or 15 or 20 | 6 × 6 × 10 | 0.36 | 4 | 2:08 |
| Semi-LASER | Symmetric EPSI | Patient | 2000 | 100 × 100 | 16 × 16 | 15 | 6 × 6 × 10 | 0.36 | 4 | 2:08 |
| *PE: phase encoding; EPSI: Echo planar spectroscopic imaging | | | | | | |||||
Results
Figure 1 shows 2D sLASER-EPSI reconstructed spectra from the phantom, using a slab across the center of phantom. 1A) 8×8 MRSI grid corresponding to conventional phase encoding (PE) clinical CSI (1 mL) and 1B) sLASER-EPSI with a much shorter scan time (40 s compared with 4:20 min). Note the uniform excitation of major metabolites such as NAA, Cho, and Cr within the grid, similar to PE-CSI. We further optimized the spatial resolution using a 16×16 matrix with varying slice thickness; 1D) the optimized protocol had a 15 mm thickness (0.36 mL) and a scan time of approximately 1 minute.After obtaining superior results in the phantom, we optimized the reliability of EPSI measurements in a volunteer as a function of slice thickness and resulting image quality as assessed by a radiologist. Figure 2C shows the optimized protocol.
Finally, the optimized protocol was tested in a patient with a with a low-grade astrocytoma who underwent treatment before MRSI. As shown in Figure 3, NAA levels in the patient were much lower on many voxels and consistent with postoperative changes.
Discussion
Because MRSI scan times in clinical protocols are usually long, there is a need for rapid imaging methods. Additionally, at higher fields like 3T, chemical shift displacements errors are higher and need to be corrected. Our study shows that MRSI acquisition with adiabatic pulses and EPSI spatial encoding could potentially be used to study spatial variations of metabolites or to assess response to radiation therapy treatment in patients with neurological diseases.Conclusion
Symmetric EPSI encoding along with semi-LASER volume localization can be used to image 2D/3D spatial metabolite maps within a clinically feasible time in brain cancer patients.Acknowledgements
This work was partially supported by NIH/NCI Cancer Center Support Grant (P30 CA008748), BTC foundation, MSKCC society grant, and B*Cured foundation
References
Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. Short echo time 1H‐MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59:1‐6.
Tkác I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41:649‐656.
Shankar RV, Chang JC, Hu HH, and Kodibagkar VD. Fast data acquisition techniques in magnetic resonance spectroscopic imaging. NMR Biomedicine 2019:32:e4046.
Coello E, Noeske R, Burns BL, et al. High-resolution echo-planar spectroscopic imaging at ultra-high field. NMR Biomedicine 2018:31:e3950
Figures
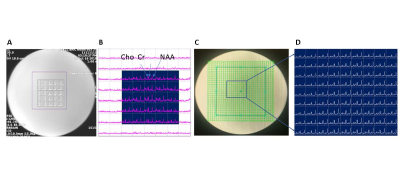
Figure 1: T1 MR images overlaid with the MRSI grid and chemical shift images. (A) conventional phase encoding PRESS spectra within the grid with its corresponding semiLASER EPSI shown in (B) obtained within a scan time of 40 s. EPSI was further tested with a higher matrix size of 16×16 (C & D). Uniform excitation of major metabolites was found to be possible within a rapid scan time.
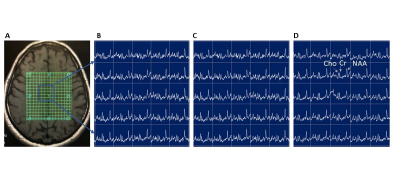
Figure 2: Optimization of metabolite signal quality for clinical assessment. A) T1 MRI with MRSI grid overlay; Rapid SemiLASER-EPSI spectra of metabolites from a slab with increasing slice thickness of (B) 10 mm, (C) 15 mm, and (D) 20 mm. Slice thickness of 15 mm was found to be best for clinical assessment.
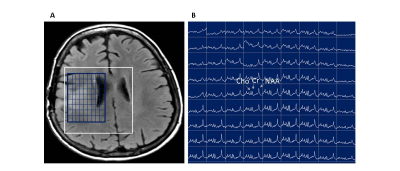
Figure 3: MRSI of a patient with brain tumor. A) T1 MRI with MRSI grid overlay; Rapid SemiLASER-EPSI spectra of metabolites from a slab with 15 mm slice thickness and a voxel size of 0.36 mL.