2870
Assessment of partial volume effects in MRS voxels applying an Altas-based brain segmentation to high-resolution 3D MRI of rat brain1The Catholic University of Korea, Seoul, Republic of Korea, 2Gacheon University, Incheon, Republic of Korea
Synopsis
The aim of this study was to apply the atlas-based brain segmentation to high-resolution 3D MRI of the mice brain, to assess a partial volume effect in the voxel. The altas-based segmentation successfully decomposed high-resolution 3D MRI into various anatomical compartments, and by volumetrically analyzing binary masks of the tissue compartment and voxels, a true contribution of the intended tissue compartment in the localized voxel can be assessed. By analyzing the metabolite-specific volume masks, an agreement of the localization between metabolites was evaluated. By evaluating the true contribution of the intended compartment and metabolite-specific agreement, localization reliability can be improved.
Introduction
Previous studies have reported that a registration of the high-resolution MRI to a brain atlas would potentialize an advanced automatic tissue compartmentalization, which can decompose MRI scans into various anatomical regions, instead a simple decomposition into white matter, gray matter, and cerebrospinal fluids (CSF). The aim of this study was to apply the atlas-based brain segmentation to high-resolution 3D MRI of the mice brain, to assess a partial volume effect and water content in the voxel. In addition, a localization reliability was evaluated by assessing a true contribution of the intended tissue compartments within the localized MRS voxels in conjugate with the chemical shift displacement for major metabolites.Materials and Methods
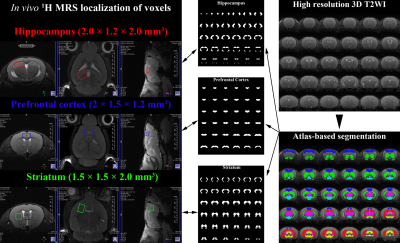
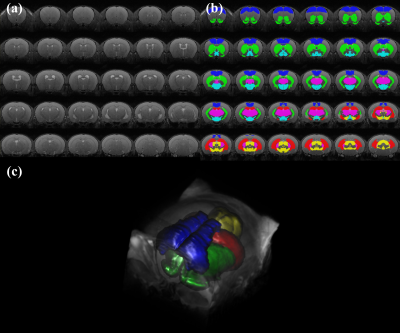
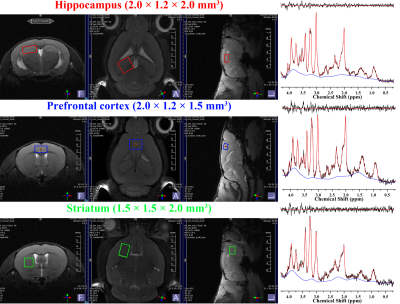
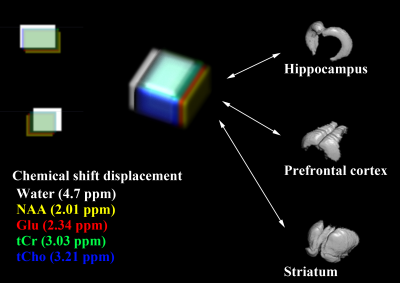
All MRI/MRS scans of five male C57BL/6J mice were acquired with Bruker BioSpec 94/20 USR system with a 72-mm linear transmit resonator, and 4-channel phased-array surface coil. High-resolution 3D T2-weighted images were acquired using TurboRARE sequence with the following parameters: TR/TEeff = 2000/33 ms; RARE factor = 16; averages = 2; field of view = 12 × 12 × 15.6 mm²; matrix size = 120 ×120 × 156; resolution = isotropic 100 μm. For the brain tissue compartmentalization, the Atlas Normalization Toolbox using ELASTIX (ANTX)-toolbox was used with the acquired MRI. Voxels of MRS were located mainly in the prefrontal cortex (PFC, 2 × 1.5 × 1.2 mm3), right hippocampus (2 × 1.2 × 2.0 mm3), and right striatum (1.5 × 1.5 × 2 mm3). The localized voxels were automatically shimmed with the Localized_shim protocol to achieve an unsuppressed water linewidth to 15–18 Hz. Point-resolved spectroscopy with a calculated-90° excitation pulse (8400 Hz; 0.5 ms) and calculated-180° refocusing pulses (3400 Hz; 1.0 ms) were used with the followings: TR/TE = 4000/15.016 ms; complex data points = 2048; average = 16 × 40; spectral BW = 5000 Hz. Before the quantification of MRS scans, an apodization with 2 Hz exponential filter was applied to each FIDs of MRS spectra, and a time-domain spectral registration was applied to the 40 sets of the apodized FIDs by using the FID Appliance (FID-A) open-source software package. The processed spectrum with total 640 averages was quantified by the LCModel with a parametrically matched basis-set simulated by the FID-A. As described in in the schematic diagram of Figure 1, a volumetric analysis was performed. After the tissue compartmentalization, binary masks of the tissue compartment label were obtained for hippocampus, PFC and striatum. Additionally, binary volume masks spatially matched with the position and size of the three MRS voxels were obtained, and also metabolite-specific volume masks for methyl-NAA (2.01 ppm), methyl-Cr (3.03 ppm), tri-methyl-Cho (3.21 ppm), C4-Glu (2.34 ppm) were created by calculating the chemical shift displacements in conjugate with the used RF pulse bandwidth, direction, and chemical shifts. Volumetric analyses between the tissue compartment label masks and voxel and metabolite-specific volume masks were performed to evaluate the localization reliability for the voxel.Results
Figure 2 illustrates representative high-resolution 3D T2-weight images, acquired in the mouse brain. As visually inspected, the acquired MRI scans showed that the SNR was generally maintained sufficient for the whole brain region with the high spatial resolution (100 μm). The co-registration and tissue compartmentalization in conjugate with the brain altas was reliably performed without any artifacts. Figure 3 illustrates representative MRS scans obtained in the (a) hippocampus, (b) prefrontal cortex, and (c) striatum in conjugate with the location of their voxels, respectively. Table 1 lists the mean concentrations and CRLB values of the three voxels. The acquired MRS spectrum showed sufficient SNR and comparable linewidth to the previous studies in the visual inspection. In addition, the CRLB values of the major metabolites seemed comparable, suggesting that high-field and short-TE MRS was reliably acquired and processed in the three voxels. Figure 4 illustrates binary masks of hippocampus, PFC, and striatum spatially matched with the voxel of MRS and each tissue compartment labels obtained from the tissue compartmentalization. In addition, the metabolite-specific volume masks were overlaid with each voxel volume mask.Discussion and Conclusion
Previous studies, which investigated the partial volume effects for the MRS, have determined the brain water content in the voxel by decomposing the whole brain simply into white matter, gray matter, and CSF, for the internal referencing. Because single voxel MRS acquires signal within the entire voxel, heterogeneous tissue compartments within the voxel would disturb interpreting the quantification results and making a correct decision. In this study, the atlas-based automatic brain segmentation was applied to high-resolution 3D MRI of the mouse brain, and the brain was decomposed into various anatomical compartments, for the advanced assessment of partial volume effects within the localized voxels of MRS. The altas-based segmentation successfully decomposed high-resolution 3D MRI into various anatomical compartments, and by volumetrically analyzing binary masks of the tissue compartment and voxels, the true contribution of the intended tissue compartment in the localized voxel can be assessed. In addition, by analyzing the metabolite-specific volume masks, an agreement of the localization between major metabolites was evaluated in conjugate with the chemical shifts displacements. By evaluating the true contribution of the intended compartment and metabolite-specific agreement of localization, localization reliability can be improved.Acknowledgements
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (2018R1A2B2005343), and the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT & Future Planning (2017M3C7A1043838).References
1. Quadrelli S, Mountford C, Ramadan S. Hitchhiker’s Guide to Voxel Segmentation for Partial Volume Correction of In Vivo Magnetic Resonance Spectroscopy. Magn Reson Insights, 2016; 9:1-8.
2. Bai J, Trinh TL, Chuang KH, Qiu A. Atlas-based automatic mouse brain image segmentation revisited: model complexity vs. image registration. Magn Reson Imaging, 2012;30:789-98.
3. Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature, 2007;445:168-76.
Figures