2861
Spatiospectral Reconstruction from Hybrid FID/SE J-Resolved MRSI Data with Limited Coverage of (k,t,tJ)-Space1Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 4Med-X Research Institute, Shanghai Jiao Tong University, Shanghai, China
Synopsis
J-resolved MRSI using hybrid FID/SE signals has recently been proposed as an effective approach to achieving rapid, high-resolution mapping of brain metabolites and neurotransmitters, an elusive goal of the MRSI community. The new data acquisition scheme poses new problems for data processing, from removal of nuisance signals to reconstruction of spatiospectral distributions. This paper presents an effective method to address these problems, utilizing a union-of-subspaces framework to absorb complementary and prior information. The proposed method has been evaluated using experimental data, producing high-quality spatiospectral distributions of metabolites and neurotransmitters from hybrid FID/SE J-resolved MRSI data with limited coverage of (k,t,tJ)-space.
Introduction
In conventional J-resolved MRSI experiments, we acquire a set of spin echoes with different TEs, which “fully” encode spatial, spectral and J-coupling information. Let $$$\rho(\boldsymbol{x},f,J)$$$ be the desired J-resolved spectroscopic imaging function, where $$$f$$$ and $$$J$$$ denote chemical shift and J-coupling frequencies respectively. The measured data $$$d(\boldsymbol{k},t,t_J)$$$ can be expressed as:$$d(\boldsymbol{k},t,t_J)=\iiint\!\rho(\boldsymbol{x},f,J)e^{-i2\pi\gamma\Delta B_0(\boldsymbol{x})t}e^{-i2\pi(\boldsymbol{k}\cdot\boldsymbol{x}+ft+Jt_J)}\,\mathrm{d}\boldsymbol{x}\,\mathrm{d}f\,\mathrm{d}J+\xi(\boldsymbol{k},t,t_J).$$Such a data acquisition scheme requires a long data acquisition time to cover the high-dimensional $$$(\boldsymbol{k},t,t_J)$$$-space. To address this problem, a new data acquisition scheme using hybrid FID/SE acquisitions with very limited coverage of $$$(\boldsymbol{k},t,t_J)$$$-space has been proposed. This acquisition scheme provides the following data:
$$$\quad$$$FID data: $$$d_{\rm{FID}}(\boldsymbol{k},t,t_J),~\boldsymbol{k}\in\mathcal{K}_{\mathrm{FID}},~t_{J}=0$$$.
$$$\quad$$$SE data: $$$d_{\rm{SE}}(\boldsymbol{k},t,t_J),~\boldsymbol{k}\in\mathcal{K}_{\mathrm{SE}},~t_{J}\in\{\mathrm{TE}_1,\mathrm{TE}_2\}$$$.
Note that the FID data were collected with ultrashort TE, short TR, large k-space coverage and no water or lipid suppression while the SE data were acquired with relatively long TR and limited k-space coverage for only two different TEs.
This acquisition poses two new data processing problems: a) removal of nuisance signals from the SE data with limited k-space coverage, and b) reconstruction of spatiospectral functions from the hybrid limited and noisy data. This paper presents an effective method to address these two problems, utilizing a union-of-subspaces framework to absorb complementary and prior spectral information. In vivo experimental results showed that the proposed method can effectively handle the overwhelming nuisance signals and reconstruct high-quality neurometabolic images using the hybrid data jointly.
Methods
We represent the desired high-dimensional J-resolved MRSI signal using the union-of-subspaces model1:$$\rho(\boldsymbol{x},f,J)=\sum_{m=1}^{M}\sum_{\ell=1}^{L_m}c_{m,\ell}(\boldsymbol{x})v_{m,\ell}(f,J),$$where $$$\{v_{m,\ell}(f,J)\}_{\ell=1}^{L_m}$$$ is a set of joint basis functions for the $$$m$$$th molecule, which can be pre-determined using quantum mechanical simulations and training data as in previous works2,3, and $$$\{c_{m,\ell}(\boldsymbol{x})\}_{\ell=1}^{L_m}$$$ are the corresponding spatial coefficients. In practice, $$$\rho(\boldsymbol{x},f,J)$$$ is composed of signals from a small number of molecules (e.g., water, lipid, metabolites and neurotransmitters); therefore, the union-of-subspaces model can effectively reduce the number of degrees-of-freedom for representing the desired spatiospectral distributions4.Nuisance removal: The FID data set has large k-space coverage and its water and lipid signals can be removed using the existing method4. To remove the lipid and water signals from the SE data with limited k-space coverage, we used the water and lipid signals from the FID data as a reference, then compensated the difference between the FID data and the SE data using the union-of-subspaces model and the generalized series (GS) model (including B0 field drift)4-6. After that, the compensated high-resolution reference signals were used to remove the lipid and water signals from the SE data.
Spatiospectral reconstruction: After nuisance removal and proper compensation for steady-state signal difference between the FID and SE data sets, the desired spatiospectral function was reconstructed using all the data jointly by solving the following optimization problem for spatial coefficients $$$C$$$7,8:$$\hat{C}=\arg\min_{C}\sum_{t_J\in\{0,\mathrm{TE}_1,\mathrm{TE}_2\}}\left\|d_{t_J}-\Omega_{t_J}\mathcal{F}\left(V_{t_J}C\right)\right\|_2^2+R(C),$$where $$$d_{t_J}$$$ denotes the nuisance removed data at $$$t_J$$$, $$$V_{t_J}$$$ and $$$\Omega_{t_J}$$$ the corresponding basis functions and sampling mask, $$$\cal{F}$$$ the imaging operator, and $$$R(\cdot)$$$ some edge-preserving regularization functional.
Results
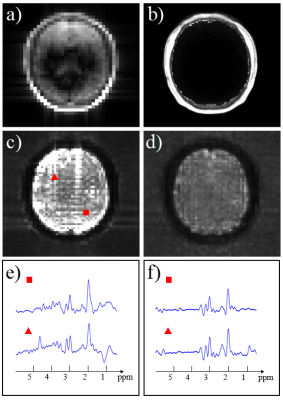
The proposed method has been evaluated using experimental data acquired from a healthy volunteer on a 3T Siemens Prisma scanner, with the following parameters: FOV = 240×240×72 mm3, TRFID = 160 ms, TRSE = 800 ms, TESE,1 = 20 ms, TESE,2 = 140 ms, FID matrix size = 78×110×24, SE matrix size = 40×40×16 and total acquisition time = 14.6 min.Results of the proposed nuisance removal scheme are shown in Fig. 1. As can be seen, the low-resolution non-lipid-suppressed SE data suffer from severe lipid contamination, and metabolites signals are overwhelmed by residual lipids even after Papoulis-Gerchberg (PG) algorithm based lipid removal9. Some water residues are also observable in the spectra. With the help of the high-resolution references from the FID dataset, the nuisance signals in the SE datasets were reduced to a negligible level.
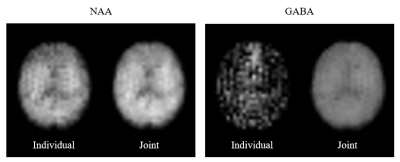
A comparison of the joint reconstruction results and individual reconstruction results is shown in Fig. 2. By comparing NAA maps, we can see a slight improvement in SNR by including more data in the joint reconstruction model. As for GABA maps, however, individual reconstruction failed to provide a meaningful estimation of GABA due to the limited SNR and spectral overlap with other resonances, while joint reconstruction allowed us to exploit the J-coupling information encoded in the data and produced much better results.
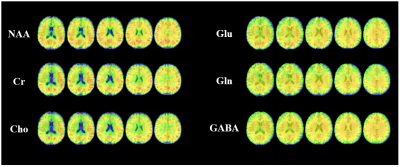
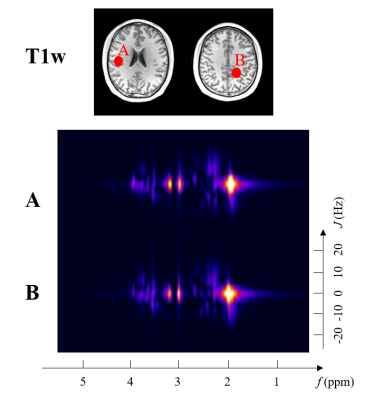
A summary of reconstructed 3D neurometabolic concentration maps is shown in Fig. 3, and a set of high-quality 2D spectra is provided in Fig. 4. As can be seen, the proposed joint reconstruction method can efficiently and effectively handle the limited $$$(\boldsymbol{k},t,t_J)$$$-space data, producing high-quality neurometabolic maps.
Conclusions
This paper addresses the data processing problems associated with rapid high-resolution J-resolved MRSI experiments with limited coverage of $$$(\boldsymbol{k},t,t_J)$$$-space. A new technique was proposed for effective removal of water and lipid signals and spatiospectral reconstruction using the FID/SE J-resolved data jointly. Experimental results show that the proposed method can produce high-quality spatial maps of metabolites and neurotransmitters.Acknowledgements
This work reported in this paper was supported, in part, by NIH-R21-EB023413, NIH-P41-EB022544 and NIH-U01-EB026978.References
[1] Lam F, Cheng B and Liang Z-P. Accelerated J-resolved MRSI using joint subspace and sparsity constraints. In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, HI, USA, 2017. 1202.
[2] Li Y, Lam F, Clifford B and Liang Z-P. A subspace approach to spectral quantification for MR spectroscopic imaging. IEEE Trans Biomed Eng. 2017; 64:2486–2489.
[3] Lam F, Li Y, Guo R, Clifford B and Liang Z-P. Ultrafast magnetic resonance spectroscopic imaging using SPICE with learned subspaces. Magn Reson Med. 2020; 83: 377-390. https://doi.org/10.1002/mrm.27980.
[4] Ma C, Lam F, Johnson CL and Liang Z-P. Removal of nuisance signals from limited and sparse 1H MRSI data using a union-of-subspaces model. Magn Reson Med. 2016;75(2):488‐497.
[5] Liang Z-P and Lauterbur PC. A generalized series approach to MR spectroscopic imaging. IEEE Trans Med Imaging. 1991; 10(2): 132-137.
[6] Ma C, Lam F, Ning Q, Johnson CL and Liang Z-P. High‐resolution 1H‐MRSI of the brain using short‐TE SPICE. Magn Reson Med. 2017; 77(2): 467-479.
[7] Lam F and Liang ZP. A subspace approach to high-resolution spectroscopic imaging. Magn Reson Med. 2014; 71(4):1349-1357.
[8] Tang L, Zhao Y, Li Y et al. Accelerated J-resolved 1H-MRSI with limited and sparse sampling of (k, tJ)-space. In Proceedings of the 27th Annual Meeting of ISMRM, Montreal, QC, Canada, 2019. 2486.
[9] Haupt CI, Schuff N, Weiner MW and Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med. 1996; 35(5): 678-687.
Figures