2857
Comparison between Cartesian and Radial Under-sampling Schemes for Fast MRI using a Deep Cascade of Neural Network1Medical Physics Program, Duke Kunshan University, Jiangsu, China, 2Department of Radiation Oncology, Duke Univeristy, Durham, NC, United States, 3Electronic Science and Engineering, Nanjing University, Nanjing, China
Synopsis
Cartesian under-sampling scheme was commonly applied in fast MRI using a deep neural network to simulate the process of fast image acquisition, however, it might not be optimal at a high under-sampling rate. Alternatively, radial under-sampling scheme was used and its efficiency was compared against that of Cartesian under-sampling scheme for T1- or T2-weighted brain, breast, prostate and cervical MRI data at various under-sampling rates. The quantitative evaluation results demonstrated that radial under-sampling scheme could outperformed Cartesian under-sampling scheme on reducing scan time while achieving comparable or better image quality.
Purpose
Magnetic Resonance Imaging (MRI) has long been considered as an effective imaging modality for diagnosis and treatment; however, the time-consuming acquisition process might degrade the effectiveness of MRI on its clinical applications. The fast MRI method using a neural network is a feasible solution to reduce the acquisition time by under-sampling MR signals, and then generating a reconstructed image using deep learning. Most of the proposed fast MRI studies using deep neural networks focused on image reconstruction based on Cartesian under-sampling schemes. Although the Cartesian under-sampling strategy is commonly used, it may not achieve an optimal result on a high under-sampling rate. As an alternative, a radial under-sampling strategy 1 has been demonstrated its potential in fast MRI 2. In this work, the radial under-sampling strategy was compared against the Cartesian under-sampling strategy for fast MRI using a deep cascade of neural network.Methods
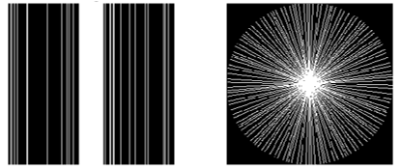
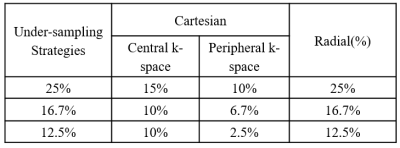
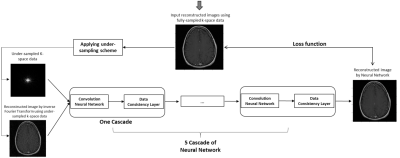
T1- or T2-weighted brain, breast, prostate and cervical MRI data from the TICA archive 3 were included in the evaluation. In the study, the under-sampled k-space data were retrospectively sparsely sampled from the full k-space data using Cartesian under-sampling and radial under-sampling schemes at 25%, 16.7% and 12.5% under-sampling rates, simulating accelerated image acquisition by factors of 4, 6 and 8. More specifically, 25% Cartesian under-sampling scheme fully sampled central k-space covering 15% k-space and randomly sampled in the rest peripheral region for the rest 10% k-space data, while the 25% radial under-sampling scheme would sparsely sample 25% k-space in a star pattern based on a golden-angle strategy, as shown in Figure 1. Similarly, the 16.7% and 12.5% under-sampling schemes were generated as illustrated in Figure 2. In this work, a deep cascade of neural network architecture (DC-CNN) 4 based on Theano framework backend with Lasagne, as shown in Figure 3, was used for improving the image quality of reconstructed image by inverse Fourier Transform using under-sampled k-space data. To increase available data, image augmentation was performed. As a strategy to improve the neural network efficiency, the neural network was trained only with T2-weighted MRI data for each anatomic site both in frequency domain using under-sampled k-space data and in image domain using reconstructed images by inverse Fourier Transform using under-sampled k-space data. The reconstruction model of each anatomic site was trained separately for 10 epochs. The evaluation data were not included in the training data and were reconstructed using the trained model with the least validation loss. The reconstructed images by neural network were compared against the reference images reconstructed from the full k-pace data, as quantified by total relative error (TRE) and mean structure similarity (MSSIM) 5. All the imaging processing and trainings of reconstruction models were carried out on a GeForce GTX 1080 card and Intel Core i7-8550U CPU card.Results
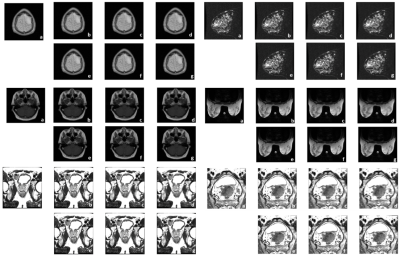
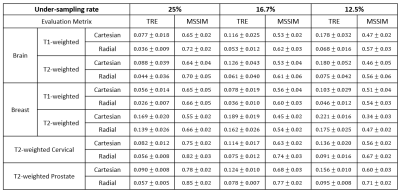
For each disease site, the training took about 2~3 hours for 10 epochs, while the reconstruction time for evaluation data only took 0.1~0.2 second per slice. Selected reconstructed images by neural network were illustrated in Figure 4. As compared against the reference images, TRE and MSSIM values of the reconstructed images were summarized in Figure 5. As indicated in Figure 5, the reconstructed images using radial under-sampling scheme consistently demonstrated smaller TRE and higher MSSIM than those using Cartesian under-sampling scheme at all 25%, 16.7% and 12.5% under-sampling rates for T1- or T2-weighted brain, breast, prostate and cervical data.Discussion
The golden-angle radial under-sampling scheme outperformed the Cartesian under-sampling scheme on various under-sampling rates. Although demonstrated with only four clinical sites, the radial under-sampling scheme could be a general strategy to achieve high quality images while greatly reducing acquisition time. For further study, the golden-angle radial under-sampling scheme could be extended to other anatomical sites such as lung, thyroid, spinal cord and abdomen as well as other MRI applications such as diffusion-weighted image (Acknowledgements
This research project could not be completed without the data collection published at The Cancer Imaging Archive (TCIA).
References
1. P. C. Lauterbur, "Image formation by induced local interactions. Examples employing nuclear magnetic resonance," Clinical Orthopaedics and Related Research, (244), pp. 3-6, 1989.
2. R. W. Chan et al, "The influence of radial undersampling schemes on compressed sensing reconstruction in breast MRI," Magnetic Resonance in Medicine, vol. 67, (2), pp. 363-377, 2012.
3. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: 10.1007/s10278-013-9622-7.
4. J. Schlemper et al, "A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction," IEEE Transactions on Medical Imaging, vol. 37, (2), pp. 491-503, 2018.
5. B. Kumar, S. B. Kumar and C. Kumar, "Development of improved SSIM quality index for compressed medical images," 2013 IEEE Second International Conference on Image Information Processing (ICIIP-2013), Shimla, 2013, pp. 251-255.
Figures