2692
Discovering MRI Imaging Biomarkers Associated with Progression of Knee Pain Using Deep Learning for Survival Analysis1Department of Radiology and Biomedical Engineering, UCSF, San Francisco, CA, United States, 2Bay Imaging Consultants, Walnut Creek, CA, United States, 3Department of Epidemiology and Biostatistics, UCSF, San Francisco, CA, United States
Synopsis
Medical attention for knee osteoarthritis (OA) is currently focused on symptomatic pain management in clinical setting. Associating future knee OA pain development with baseline characteristics of knee OA cohort is important in understanding of disease prognosis and designing treatment strategies. We built a predictive model of knee OA pain-free survival time using baseline MRI and other image-independent clinical measurements.
INTRODUCTION
Knee osteoarthritis (OA) is one of the leading causes of chronic disability in the US. Without treatments available to reverse the progression of structural deterioration except for the surgical intervention, currently, medical care for knee OA is focused on symptomatic pain management as pain is the most prominent and disabling symptom of knee OA. Understanding future knee pain development patterns and associating them with baseline characteristics of the cohort would help to improve our understanding of disease prognosis and designing treatment strategies. In this study, we model knee pain progression as individual survival outcomes, time to develop knee pain for a subject who currently does not have pain.METHODS
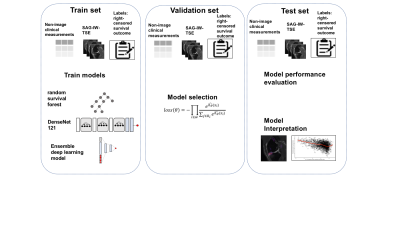
Study populationDataset for this study was obtained from the longitudinal study conducted by Osteoarthritis Initiative (OAI). We selected participants who reported no problems in pain according to either Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) or Knee Injury and Osteoarthritis Outcome Scores (KOOS) pain scores at the enrollment visit (Figure1)1, 2. Each subject’s KOOS pain scores reported in every follow-up visit from 0 to 96 months were considered for overall survival and censoring information for its relative granularity. The data were randomly split into a training- (n=1,592), a validation-(n=391), and test set (n=485).
Survival outcomes
We defined the encounter as the decrease in KOOS pain score greater than minimum detectable changes (MDC) in knee pain, 13 KOOS points. Follow-up for each subject varied. It began at the date first image was acquired. The endpoint of survival time was calculated from the date of first MRI scan to the date they qualified as having the outcome of interest. All other participants were censored at the last follow-up date or end of the study.
MR imaging
Sagittal Intermediate-Weighted fat-saturated Turbo Spin Echo sequences in Digital Imaging and Communications in Medicine format were obtained from OAI. The imaging parameters were: TE / TR = 30 / 3,200ms, FOV = 16x16cm2, in-plane spatial resolution = 0.357 x 0.357 mm2, matrix size = 448 x 448, slice thickness = 3mm, 37 slices, bandwidth = 248 Hz/pixel, refocusing flip angle = 180.
Deep learning architecture and training details
Our DL architecture was based on DenseNet1213. We modified the convolutional layers, batch normalization layers, pooling layers, and leaky rectified linear unit layers for 3D volumetric image input4, 5. The last fully connected (FC) layer has the output of dimension one, which yields the log hazard ratio. We used negative partial log-likelihood as the loss function. To combine heterogeneous multi-scale datasets, we followed an ensemble approach similar to synthetic forest regression6.
Model evaluation and interpretation
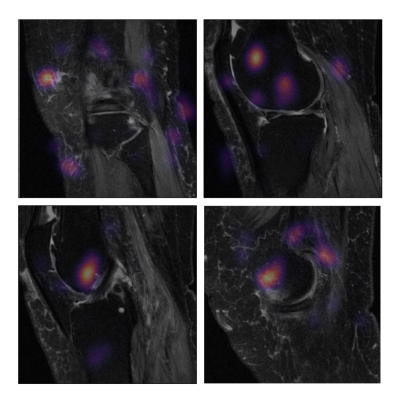
We provide the prediction error rate, 1 - Harrell’s concordance index, evaluated on the hold-out test set. To assess the variation of the estimates, we provide 95% confidence intervals (CI) generated from bootstrapping principle (B = 1,000) on the hold-out test set. We provide occlusion maps for one hundred MRI from the hold-out test set that were predicted to have the highest partial log hazard rate. The pixels associated with the top 10% decrease were identified as image biomarkers potentially linked to shorter pain-free survival time. The overview of proposed pipeline is shown in Figure2.
RESULTS
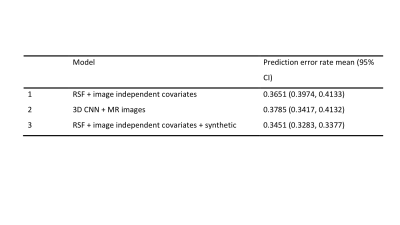
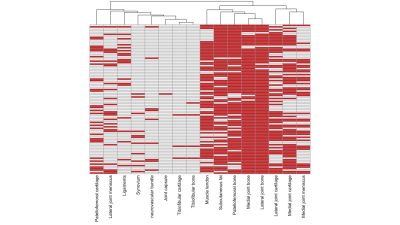
The prediction error rates of random survival forest model were 0.3683 (95% CI: 0.3243, 0.4142). The 3D CNN model using the MRI only as input achieved the prediction error rates of 0.3785 (95% CI: 0.3417 – 0.4132). The clincal and image ensemble DL model had the lowest prediction error rate of 0.2840 (0.2602, 0.3242) (Figure3). The occlusion maps for those who were predicted the highest risk of knee OA pain development highlighted lateral-, medial -, and patellofemoral- joint bones (96%, 94%, and 83%, respectively) (Figure4). Lateral-, medial-, and patellofemoral- joint cartilage were also prevalent (60%, 63%, 32%) (Figure5).DISCUSSION
A major strength of this study is a complete data-driven analysis for predicting time to knee OA pain development by utilizing DL models and large datasets. As OA is a multi-faceted disease, a systemic integrative view is required in combining multi-modal datasets with heterogeneous scale and dimension. The limitations of this study should be noted. We made strong assumptions that the behavioral or clinical interventions happened during the study period did not affect the trend of pain progression and that drop-out or missing data points are not informative, but random.CONCLUSION
To our knowledge, this is the first study to examine the association between features visualized in knee MRI and the right-censored survival outcome of knee pain development. Our ensemble model provides predicted risk of developing knee pain for individuals given complex and heterogeneous datasets, knee MR along with clinical measurements acquired at baseline.Acknowledgements
- This study was funded by the National Institutes of Health - National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH-NIAMS). Grant numbers: R00AR070902 (VP), R61AR073552 (SM/VP)
- The OAI is a public-private partnership comprised of five contracts (N01- AR-2-2258; N01-AR-2-2259; N01-AR-2- 2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health.
References
1. Roos, E.M. and L.S. Lohmander, The Knee injury and Osteoarthritis Outcome Score (KOOS): from joint injury to osteoarthritis. Health and quality of life outcomes, 2003. 1(1): p. 64.
2. Bellamy, N., et al., Validation study of WOMAC: a health status instrument for measuring clinically important patient relevant outcomes to antirheumatic drug therapy in patients with osteoarthritis of the hip or knee. The Journal of rheumatology, 1988. 15(12): p. 1833-1840.
3. Huang, G., et al. Densely connected convolutional networks. in Proceedings of the IEEE conference on computer vision and pattern recognition. 2017.
4. Faraggi, D. and R. Simon, A neural network model for survival data. Statistics in medicine, 1995. 14(1): p. 73-82.
5. Katzman, J.L., et al., DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC medical research methodology, 2018. 18(1): p. 24.
6. Ishwaran, H. and J.D. Malley, Synthetic learning machines. BioData mining, 2014. 7(1): p. 28.
Figures
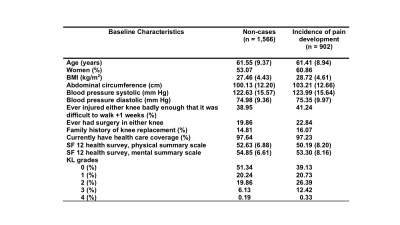
- Among study cohorts, we selected 2,468 participants who reported no problem in pain according to the Knee Injury and Osteoarthritis Outcome Scores (KOOS) or Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) pain scores.
- When KL grades are missing, it was predicted by a deep learning model developed in-house.
- SF = short form; BMI = Body mass index; KL = Kellgren-Lawrence