2668
Deep learning based fully automated screening pipeline for abnormal bone density using a short lumbar Dixon sequence1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Radiology, The Third Affiliated Hospital of Southern Medical University (Orthopaedic Hospital of Guangdong Province), Guangzhou, China, 3Radiology, Stony Brook Medicine, Stony Brook, NY, United States, 4Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States
Synopsis
Bone marrow fat fraction (BMFF) has been recognized as one of the quantitative image biomarkers to identify abnormal bone density using modified Dixon sequence. However, this method requires manual segmentation which limits its adoption in clinical practice. In this study, we developed a fully automated radiomics pipeline using deep learning based segmentation and validated its performance comparable to manual segmentation. This finding will facilitate the clinical utility of the entire pipeline as a screening tool for early detection of abnormal bone density.
Introduction
Osteoporosis has become a major public health problem worldwide; after 50 years of age, the risk of osteoporotic fractures is 50% for women and 20% for men among many Western populations[1]. Therefore, it is critically important to identify patients at high risk of osteoporosis at an early stage. A previous study[2] demonstrated the capability of bone marrow fat fraction (BMFF) extracted using Modified Dixon (mDixon) Quant to predict abnormal bone density, however, the clinical utility is limited by the requirement of manual segmentation, and the predictive power of more quantitative imaging biomarkers remains to be explored. In this study, we demonstrate a fully automated end-to-end radiomics pipeline using reliable segmentation from uNet[3] convolutional neural network (CNN) architecture.Materials and Methods
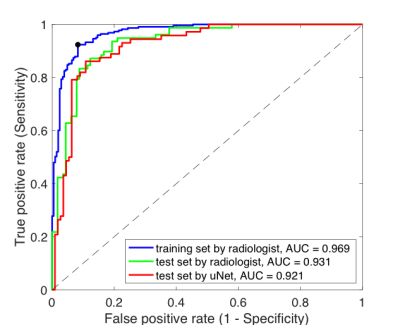
Population and image acquisition: A total of 257 participants without known spinal tumor, history of trauma, dysplasia, spinal surgery or hormone therapy were recruited in this prospective study with approval by local IRB. All patients underwent QCT examinations to obtain bone mineral density (BMD) of lumbar vertebrae (L1 to L3). The recommended threshold of 120 mg/cm3 by the International Society for Clinical Densitometry (ISCD)[4] was used to determine abnormal bone density. mDixon Quant with six-echo, seven fat peaks modeling was obtained from a 3.0T MR scanner (Ingenia, Phillips, Amsterdam, Netherlands) and T2* correction was then performed to quantify the vertebral fat fraction.Image processing and analysis: Rectangular regions-of-interest (ROIs) were delineated in mid-sagittal view of each vertebral body on the fat fraction maps by a radiologist with 12 years of experience. The dataset was temporally divided into two sets (Training set n = 142 with 74 in abnormal bone density class, and test set n = 64 with 26 in abnormal bone density class) to develop radiomics signature. uNet CNN was developed on the same training set to segment L1-L3 from fat fraction maps. A total of 92 radiomic features were extracted using in-house Matlab software[5], including first-order, GLCM and GLRLM features from both fat fraction and T2* maps. Feature pre-selection was performed to identify features with high reproducibility and predictive power (Intraclass correlation coefficient > 0.5, Mann Whitney U-test p<0.05, Spearman correlation |ρ|<0.8). The final prediction model was built via least absolute shrinkage and selection operator (LASSO) and logistic regression with 3-fold cross-validation. The receiver operating characteristic (ROC) analysis was performed to evaluate the model performance on the test dataset with ROIs drawn by the radiologist and uNet.
Results and Discussion
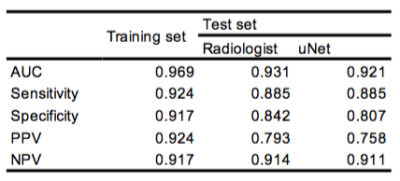
As shown in Figure 1, uNet achieved a mean Dice coefficient of 0.912±0.062 in the test set. In terms of the performance in predicting abnormal bone density, the CNN segmentation achieved AUC (0.921) comparable to manual segmentation (0.931) for the radiomics-based prediction model. This radiomic model achieved a superior performance compared to previously model based only on BMFF and clinical variables [2]. In particular, the model achieved excellent NPV at 0.914/0.911 for manual and uNet segmentation respectively, which is clinical important to identify patients with normal bone density as a screening tool. Receiver operating characteristic data are shown in Figure 2 and prediction metrics are summarized in Table 1.Conclusion
A fully automated radiomics pipeline using reliable segmentation from CNN architecture was developed and validated with comparable performance to manual segmentation. This finding will help advance the clinical adoption of the entire pipeline as a screening tool for early detection of abnormal bone density.Acknowledgements
No acknowledgement found.References
1. Liu, J., et al., State of the art in osteoporosis risk assessment and treatment. J Endocrinol Invest, 2019. 42(10): p. 1149-1164.
2. Zhao, Y., et al., Prediction of Abnormal Bone Density and Osteoporosis From Lumbar Spine MR Using Modified Dixon Quant in 257 Subjects With Quantitative Computed Tomography as Reference. Journal of Magnetic Resonance Imaging, 2019. 49(2): p. 390-399.
3. Ronneberger, O., P. Fischer, and T. Brox. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015. Cham: Springer International Publishing.
4. Engelke, K., et al., Clinical use of quantitative computed tomography and peripheral quantitative computed tomography in the management of osteoporosis in adults: the 2007 ISCD Official Positions. J Clin Densitom, 2008. 11(1): p. 123-62.
5. Renee Cattell, S.C., Chuan Huang, Robustness of Radiomic Features in MRI: Review and A Phantom Study. Visual Computing for Industry, Biomedicine, and Art, 2019.