2664
Knee MR Image Translation from Multi Contrast Images for Fat Suppression via Deep Learning with Perceptual Loss1Yonsei University, Seoul, Republic of Korea, 2University of California-San Diego, San Diego, CA, United States, 3VA San Diego Healthcare System, San Diego, CA, United States
Synopsis
The goal of this study is to generate PD Fat Suppression (PD FS) knee magnetic resonance (MR) image using deep neural network with three multi-contrast MR images (T1-weighted, T2-weighted, PD-weighted images). The results of our study show that our deep learning model can learn the relations between the input multi-contrast images and PD FS image and demonstrate the feasibility of deep learning can generate the features which have an effect on diagnosis.
Purpose
In a typical knee MR protocol, proton density (PD) or T2-weighted (T2) fat suppressed (FS) images are acquired not only for soft tissue and synovial fluid contrast1, but also for assessment of bone marrow edema.2-3 T1-weighted (T1) images without FS provide high marrow signal and contrasting low signal from the bone. It would be useful (i.e., time savings, or in case FS images are not available) if FS images could be generated from other existing images. The purpose of this study was to demonstrate feasibility of deep learning algorithm to generate PD FS images from a set of T1, T2, and PD images without FS.Method
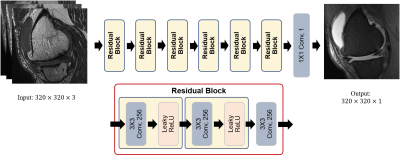
We trained a deep learning model that uses multi-contrast images to generate a synthetic FS image mimicking the ground truth PD FS image. The model is a fully convolutional neural network that accepts a stack of three T1, T2, and PD images, and outputs an image that is compared with the ground truth PD FS image to train the model through back propagation. The overall architecture of our network is shown in Fig. 1. All the image data used in this study have been aligned using rigid body registration prior to training. We used four cost functions for model optimization. The first cost function is voxel-wise mean squared error function. The second cost function is patch-based variation error function. The third cost function is registration error compensation function. And the last one we used is the perceptual loss function based on VGG-16 network pretrained with Imagenet.4We obtained anonymized MR data of 8 knees with varying pathology, imaged with four different spin echo sequences (Fig. 2ABCD): T1 (TR~650 ms, TE~20), T2 (TR~1500 ms, TE~70 ms), PD (TR~3900, TE~40), and PD FS. 127 position-matched slices (20 had bone marrow edema, 33 had moderate to severe effusion) were sorted and prepared as 3-ch (one for each of T1, T2, and PD image) input and 1-ch ground truth (PD FS). Of these, 22 slices were randomly selected for validation. DL image translation based on a linear architecture with 7 residual blocks which consist of three 3x3 convolutional layers and two nonlinear layers (Leaky ReLU), Our model was implemented using GPU tensorflow library and trained with Adam optimizer.
Result and Discussion
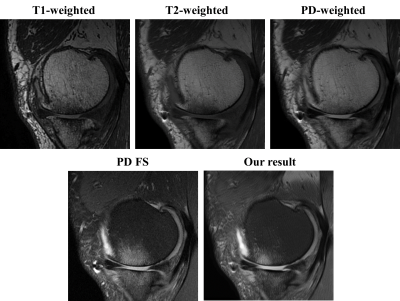
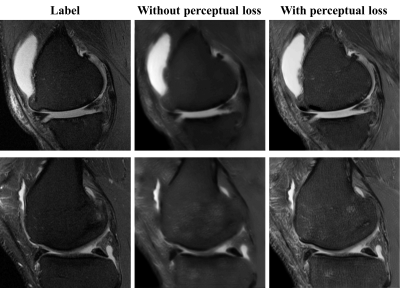
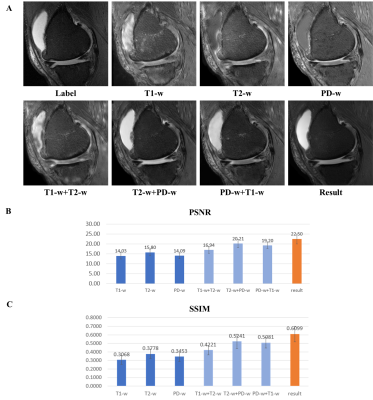
Our model is trained for about five hours and yielded visually acceptable results (Fig.2). Compared to ground truth PD FS image, our result images accurately depicted areas of bone marrow edema and synovial fluid with high signal intensity. Figure 3 shows comparisons between two cases of our results. For one case, the perceptual loss function was not used, and the second case, the perceptual loss function was used for the training. When the perceptual function is used, it can be seen that the blurring artifacts in the image are considerably reduced. This shows that the perceptual loss function helps to learn the sharpness of the original image. Figure 4 shows how the results vary according to the input combinations. As shown in Fig. 4A, when only one input is used, the contrast of the label PD FS images is not properly expressed due to the lack of information. When two contrast images are used as inputs, the contrast expression is improved compared to the results based on one input. Among them, the inclusion of PD images yields better results than the cases trained without PD images. This indicates that PD image plays the most important role in generating PD FS image. Fig. 4BC shows these by PSNR and SSIM scores, respectively. The best case for both metrics is the case that all three inputs are used. When two inputs are used, the cases including PD images achieved higher scores than the case without PD images.Conclusion
This study demonstrates the feasibility of deep learning image translation in generating fat suppressed images depicting features of clinical importance. Future refinement may include denser network, robust loss function, and increased training data size and types.Acknowledgements
This research was supported in parts by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01 AR066622 in support of Dr. Bae, and Award Number R01 AR064321 in support of Dr. Chung, as well as Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2019R1A2B5B01070488), Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2018M3A9H6081483) in support of Dr. Hwang.References
[1] Yoshioka H, Stevens K, Genovese M, Dillingham MF, Lang P. Articular cartilage of knee: normal patterns at MR imaging that mimic disease in healthy subjects and patients with osteoarthritis. Radiology. 2004;231(1):31-38.
[2] Bredella MA, Tirman PF, Peterfy CG, et al. Accuracy of T2-weighted fast spin-echo MR imaging with fat saturation in detecting cartilage defects in the knee: comparison with arthroscopy in 130 patients. American journal of roentgenology. 1999;172(4):1073-1080.
[3] Mohr A. The value of water-excitation 3D FLASH and fat-saturated PDw TSE MR imaging for detecting and grading articular cartilage lesions of the knee. Skeletal radiology. 2003;32(7):396-402.
[4] Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint. 2014;arXiv:1409.1556.
Figures