2661
Automated Cartilage Segmentation for Clinical Knee MR Images using Transfer Learning1Program of Advanced Musculoskeletal Imaging, Cleveland Clinic, Cleveland, OH, United States, 2Radiology, Cleveland Clinic, Cleveland, OH, United States
Synopsis
Laborious and time-consuming manual or semi-automatic cartilage and meniscus segmentation, which in addition suffers from intra and inter reader variability, has been one of the major hurdles of developing and applying techniques such as quantitative magnetic resonance imaging in routine clinical practice for improved osteoarthritis patient treatment and management plans. In addition, effective and robust deep learning based automatic cartilage and meniscus segmentation models are still lacking in heterogenous clinical settings. The purpose of this study is to assess the feasibility of building an automatic cartilage segmentation model using transfer learning with limited and heterogenous clinical MR scans.
Introduction
Knee pain is one of the major causes of disability affecting approximately 50% of patients over the age of 50, where 20% of patients reported severe disability as a result1. The prevalence of osteoarthritis (OA) and meniscal tears in this patient population ranges from 20% - 40% with more than 90% having both conditions2. In the United States, more than $51 billion is spent annually to treat patients with OA alone. Although techniques, such as quantitative magnetic resonance (MR) imaging, have been developed for detecting early degeneration of cartilage, effective prediction and early diagnosis of cartilage degeneration and meniscal tears are still challenging in routine clinical practice, resulting in poor patient treatment and management plans. One of the major hurdles of developing and applying these models and techniques is the laborious and time-consuming manual or semi-automatic cartilage and meniscus segmentation, which in addition suffers from intra and inter reader variability. Efforts have been made to build fully automatic cartilage and meniscus segmentation models based on deep learning. These models, however, are typically trained on homogeneous research dataset, such as the osteoarthritis initiative (OAI) dataset, which cannot be directly translated into clinical routines with different MR scanners, imaging parameters, and image qualities. Moreover, the enormous amount of training data needed prohibits to train such a model from scratch using clinical data. The purpose of this study is to assess the feasibility of building an automatic cartilage segmentation model using transfer learning with limited and heterogenous clinical MR scans.Methods
The architecture of the deep learning segmentation model was based on the conditional generative adversarial networks (cGAN)3 and U-Net4. The proposed cGAN model consisted of two parts, a 6-layer convolutional discriminator and a 10-layer U-Net generator, where they evolved by competing against each other. The U-Net was used in place of the generator as it has shown good performance in segmenting tissues in knee MR images. It contained 5 encoding layers and 4 decoding layers with skip connections, and an output layer. The segmentation model was first trained on the manually segmented homogeneous OAI dataset, which contained 176 sagittal knee MR images with cartilage segmented based on the 3D sagittal double-echo steady state (DESS) sequence. Each image consisted of 160 slices (0.7mm slice thickness) with FOV 14cm and matrix size 384×384. The model was then transferred to a heterogenous clinical dataset by applying transfer learning. The clinical dataset contained 25 sagittal 2D fast spin-echo (FSE) fat-suppressed proton density weighted clinical knee MR images from 9 Cleveland Clinic sites, 6 different MR scanner models, and 2 different magnetic field strength (ten 1.5T and seven 3T), with different number of slices (25 - 40) and various matrix sizes and heterogenous image contrast and quality. Each set of MR images was manually segmented by a trained radiologist into six compartments for the articular cartilage, and then combined into femoral cartilage, medial tibial cartilage, lateral tibial cartilage, and patellar cartilage. The 25 sets of MR images were randomly divided into 20 for training and validation, and 5 for testing. The training and validation dataset was further augmented by counterclockwise 90-degree rotations and mirroring. The ADAM optimizer was used for model training with an initial learning rate of 1e-3 and a decay rate of 0.9. The batch size was set to 10. The segmentation performance was evaluated using the Dice coefficient comparing the automatic segmentation and the manual segmentation.Results
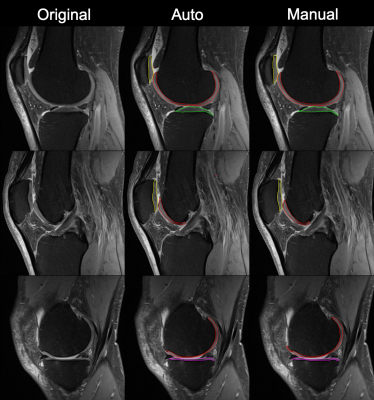
The training of the transfer learning model on the heterogeneous clinical MR images finishes in 10,000 iterations. The average Dice coefficient of applying the deep learning model on the held-out test set after transfer learning is 0.817 with standard deviation of 0.035, compared to that of 0.495 with standard deviation of 0.103 before transfer learning. Figure.1 shows a comparison between the automatic and manual cartilage segmentation on an MR image slice collected on a Siemens 3T Verio scanner, where the fist column contains the original MR images, the second column is the automatic segmentation, and the third column is the manual segmentation. The red, green, magenta, and yellow colors represent femoral, lateral tibial, medial tibial, and patellar cartilage segmentations respectively.Discussion
This study developed a promising deep learning model for automatically segmenting articular cartilage on heterogeneous image dataset with limited training data using transfer learning. The model achieved a Dice coefficient of 0.817 on a clinical dataset with various MR scanner models, field strengths, image resolution, contrast, and quality. The performance of the model can be further improved by optimizing the architecture and parameters of the model. More sophisticated image augmentation on this limited clinical MR image dataset can also help improve the model performance. The automatic segmentation model can be potentially used for important clinical applications such as quantitative MR imaging, cartilage lesion detection, and patient outcome prediction.Conclusion
The transfer learning model has showed its ability to automatically segment a wide range of clinical MR images with very small data size obtained with different scanners, different imaging parameters, and different image qualities. Fully automated segmentation of clinical knee MR images will enable the clinical application of quantitative MR imaging technique and other prediction models for improved patient treatment and management plans.Acknowledgements
No acknowledgement found.References
1. Jinks C, Jordan K, Croft P. Measuring the population impact of knee pain and disability with the Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC). Pain, 100, 1-2 (2002), 55-64.
2. Lawrence RC, Felson DT, Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum., 58, 1 (2008), 26-35.
3. Mirza M, Osindero S. Conditional Generative Adversarial Nets. arXiv (2014), arXiv:1411.1784.
4. Norman B, Pedoia V, Majumdar S. Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology, 288, 1 (2018), 177-185.
Figures