2658
A 3D Convolutional Deep Neural Network for lumbar plexus segmentation1Center for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, UCL, London, United Kingdom, 2Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, UCL, London, United Kingdom, 3Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, Brain MRI 3T Research Center, IRCCS Mondino Foundation, Pavia, Italy, NMR Research Unit, Queen Square MS Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, UCL, London, United Kingdom, 4Universitat Oberta de Catalunya, Barcelona, Spain, Center for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, NMR Research Unit, Queen Square MS Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, UCL, London, United Kingdom
Synopsis
A fully automated approach for lumbar plexus segmentation that could facilitate quantitative MRI assessments is presented. The approach is based on a 3D cascaded Convolutional Deep Neural Network (CNN) with concatenated loss function and optimized data augmentation policy. The method offers single modality segmentation and uses as input a commonly used 3D acquisition for peripheral nerve imaging. The performance analysis of the predicted segmentation results in comparison to manually segmented masks revealed 68% agreement. Future improvements in the predictive performance of the proposed method are anticipated by involving much larger datasets to reduce overfitting and improve CNN generalization ability.
Purpose
The peripheral nervous system can be affected by a wide range of pathological conditions. Whilst dedicated magnetic resonance imaging (MRI) acquisitions have recently enabled exceptional visualisation of the peripheral nerves in vivo, these methods only provide a qualitative assessment of the nerves rather than more specific information about the underlying pathophysiological mechanisms involved in each case. Quantitative MRI methods are being developed to address this gap in knowledge but are heavily relying on accurate segmentation of the peripheral nerves, which is a subjective procedure and prone to errors. In this proof of concept study, a method for fully automated segmentation of the lumbar plexus is presented, which is a 3D cascaded Convolutional Deep Neural Network known to provide more accurate segmentation of complex structures. It is worth mentioning that in comparison to standard CNN, cascaded CNN architectures1,2 are also known to reduce significantly incorrect classification, detected through hierarchy of training models.Methods
MRI dataset: A retrospective analysis of data from 9 healthy controls (4 females, mean age 36.2 years, range 29-61) 5 with scan-rescan and 4 with a single scan previously acquired using a Philips Achieva 3T system and the product 15-channel SENSE spine coil was performed. At the time of scanning, the lumbosacral spine was imaged coronally using the commonly used 3D SHINKEI sequence for optimal nerve visualisation, as follows3- 5 : TR = 2500 ms; TE = 192 ms, FOV = 180 × 180 mm2 , voxel size = 1 x 1 x 1 mm3 , number of averages = 2, TSE factor = 100, improved Motion-Sensitized Driven-Equilibrium (iMSDE) duration = 50 ms, 81 slices, scanning time of 10m20s. The imaging volume was positioned at the superior margin of the L1 vertebral body with the volume extending caudally towards the sacral segments, to ensure coverage of the L1-L5 segments in all subjects. Image analysis: For each participant, the lumbar segments L1-L5 were manually delineated by one experienced rater and a binary mask was created for each one using FSLview6 .Computational approach: The CNN model for lumbar segmentation used in this study was a cascaded model of two 3D patch-wise neural networks with identical CNN model architecture. The CNN was trained and tested on a balanced set of 3D SHINKEI images and masks composed of the same number of positive and negative samples and random selection of subjects for training and cross validation7 (see Fig 1). The main differences between the implemented CNN method and previous work7 were a concatenated loss function (made of Categorical cross entropy, Jaccard, Dice, Hausdorff distance, Precision), and additional data augmentation such as rotation and flipping images on batches. This network accepts only one modality (3D SHINKEI) as input and creates two probability masks (with regards to two models) and a final segmentation (binary mask) as output.
Network training and testing: To train and test our CNN we used 5 scan-rescan subjects for training and cross validation and 4 subjects with a single scan for testing, each subject with one 3D SHINKEI acquisition and manually segmented lumbar segment (binary mask).
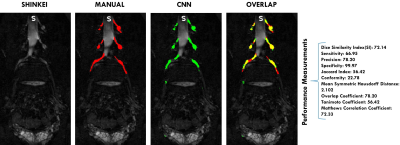
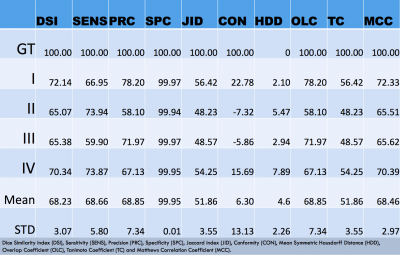
Performance analysis: The automated segmentation performance was assessed from a comparison with the manual segmentations (Ground Truth), using the following performance metrics: Dice Similarity Index (DSI), Sensitivity (SENS), Precision (PRC), Specificity (SPC), Jaccard index (JID), Conformity (CON), Mean Symmetric Hausdorff Distance (HDD), Overlap Coefficient (OLC), Tanimoto Coefficient (TC) and Matthews Correlation Coefficient (MCC).
Results
Fig 2 and Fig 3 show a visual comparison between manual and CNN-predicted segmentations, included with the performance results. Table1, instead, reports the results of the comparison between manual and CNN segmentation on the 4 testing subjects (never seen before by the CNN), where for instance Dice Similarity Index reaches its best value at 72.14 and worst score at 65.07.Discussion
In this proof of concept study, we have developed and tested a method for automated segmentation of the lumbar plexus from a standard 3D MRI acquisition protocol. We used a Convolutional Deep Neural Network implementation, which was specifically optimised for this task. The proposed fully automated image analysis method could be used in future research studies to facilitate quantitative MRI assessments and could provide an accurate and detailed knowledge of the lumbar plexus in a relatively short analysis time. Despite the modest size of the dataset used for the development of the CNN, performance results have demonstrated a high level of success; however, this study suggests that training and running CNN with much larger datasets could improve the image segmentation further.Acknowledgements
The UK MS Society and the UCL-UCLH Biomedical Research Centre for ongoing support. CGWK receives funding from ISRT, Wings for Life and the Craig H. Neilsen Foundation (the INSPIRED study), from the MS Society (#77), Wings for Life (#169111), Horizon2020 (CDS-QUAMRI, #634541). D.C.A. has received funding for this work from Engineering and Physical Sciences Research Council Grants M020533, M006093, and J020990, as well as the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreements 634541 and 666992. FP is a non-clinical Postdoctoral Guarantors of Brain fellow.References
1) Littmann E, Ritter H. Generalization abilities of cascade network architecture. In Proceedings of the 5th International Conference on Neural Information Processing Systems (NIPS'92), S. J. Hanson, J. D. Cowan, and C. L. Giles (Eds.). Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, 188-195, (1992).
2) Guo Y, Bai L, Lao S, et al. A Comparison between Artificial Neural Network and Cascade-Correlation Neural Network in Concept Classification. Springer International Publishing, ISBN="978-3-319-13168-9", 248–253, (2014).
3) Yoneyama M, Takahara T, Kwee T.C, et al. Rapid high-resolution MR neurography with a diffusionweighted pre-pulse, Magn. Reson. Med. Sci. 12(2):111–119 (2013).
4) Kasper J.M, Wadhwa V, Scott K.M, et al. SHINKEI-a novel 3D isotropic MR neurography technique: technical advantages over 3DIRTSE-based imaging, Eur. Radiol. 2015;25(6):1672–1677.nvolutional neural network approach. CoRR abs/1702.04869 (2017).
5) Yiannakas M, Schneider T, Yoneyama M, et al. Feasibility of magnetisation transfer ratio measurements in the proximal lumbar plexus using healthy volunteers at 3T. ISMRM 27th Annual Meeting & Exhibition, (2019).
6) Jenkinson M, Beckmann C, Behrens T E J, et al. FSL. NeuroImage 62, 782–790 Academic Press Inc., (2012).
7) Valverde S, Cabezas M, Roura E, et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage, 155, 159–168 (2017).
Figures