2653
Compartment-specific Knee Cartilage Segmentation using Deep Learning1Research Unit of Medical Imaging, Physics and Technology, University of Oulu, Oulu, Finland, 2Department of Diagnostic Radiology, Oulu University Hospital, Oulu, Finland
Synopsis
In this study, we developed a method for compartment-specific segmentation of knee cartilage from 3D-DESS MR images which jointly utilizes deep learning and atlas-based approaches. The method was applied to compare the performance of two deep learning-based segmentation models on two independent datasets. One of the models achieved new state-of-the-art in knee cartilage segmentation on the Osteoarthritis Initiative data and was more robust to the changes in MRI protocol. Detailed analysis performed using our method showed how the performance improvements are localized compartment-wise. The method can be used to select the most accurate segmentation model for the considered clinical problem.
Introduction
Automatic methods based on deep learning (DL) have recently achieved promising results in knee cartilage segmentation from MRI data 1,2,3. Such methods, however, are sensitive to inhomogeneities in input data, e.g., caused by changing the MRI protocol 4. These failures may remain unnoticed, since the DL-based segmentation can introduce bias within tissue compartments across the datasets, while knee MRI segmentation methods are typically assessed tissue-wise. Additionally, measured at tissue-level, segmentation metrics do not fully reflect the morphological changes of articular cartilage. Accordingly, such evaluation may prioritize the method with lower accuracy in the regions of interest, but higher in the ones that are trivial to segment. Therefore, assessment and comparison of automatic segmentation methods have to be done compartment-wise, in a clinically meaningful manner.In this study, we propose a multi-stage method for compartment-specific segmentation and analysis of knee cartilage tissues. The first stage of the pipeline employs a fast and accurate DL-based segmentation model. The second stage uses an atlas-based method for automatic compartmentalization of the segmented tissues.
Methods
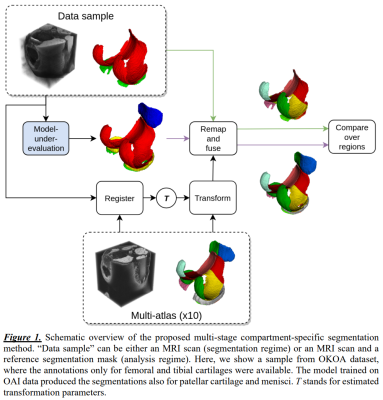
Our multi-stage method performs segmentation of the cartilage tissues and their subsequent compartmentalization (Figure 1). For training and evaluation of the segmentation models, we used a subset of data from the Osteoarthritis Initiative (OAI) 5. The dataset consisted of 176 3D-DESS MRI scans (3T Siemens MAGNETOM Trio, quadrature transmit-receive knee coil, voxel 0.37x0.37x0.7mm3, matrix 384x384, FOV 140mm, 160 slices, TR 16.3ms, TE 4.7ms, flip angle 25°) with semi-automatic annotations by iMorphics 6. The data was split patient-wise into training and test subsets, 140 and 36 scans respectively, stratified by radiographic OA severity (Kellgren-Lawrence scale). For evaluation, we additionally employed the OKOA dataset 7, which contains 44 3D-DESS scans and mainly differs in acquisition hardware and spatial resolution (3T Siemens MAGNETOM Skyra, 15-channel transmit-receive knee coil, voxel 0.59x0.59x0.6mm3, matrix 256x256, FOV 150mm, 160 slices, TR 14.1ms, TE 5ms, flip angle 25°). Annotations for femoral and tibial cartilage tissues were produced by our research group. Two DL-based methods were developed using a training subset of OAI to segment femoral, tibial, patellar cartilages, and menisci. Each method consisted of an ensemble of models trained in 5-fold cross-validation setting. As a first method (A), we used the previous state-of-the-art in the domain 3. There, a modified U-Net architecture with 6 depth levels and 24 base filters was trained from scratch. The second method (B) was based on transfer learning and used VGG19 8 backbone pre-trained on ImageNet as an encoder, otherwise being similar to A. To improve the method robustness, we used mixup regularization 9.The second stage of our approach performed compartmentalization of the tissues. We selected ten scans from the training subset of OAI to construct a multi-atlas, maintaining similar distribution of Body Mass Index as in the whole subset. The annotation of multi-atlas was done in accordance with MRI Osteoarthritis Knee Score (MOAKS) guidelines 10.
During evaluation, we segmented the scans from the test subset of OAI and whole OKOA using both methods – A and B. Next, we used rigid registration of the multi-atlas to each scan to estimate the similarity transforms. These transforms were used to warp the masks from multi-atlas to the reference and the automatic segmentations. The segmentation masks for each cartilage tissue were then divided into compartments voxel-wise based on the Euclidean proximity to the valid compartments from the atlas. Finally, we used majority voting to fuse the compartment-level maps across the multi-atlas. To quantify the segmentation accuracy, we used volumetric Dice score coefficient (DSC) computed tissue- and compartment-wise.
For training the models, we used PyTorch framework and NVIDIA RTX 2080ti graphics card. For registration, we employed elastix 11. For compartmentalization we used SciPy 12.
Results
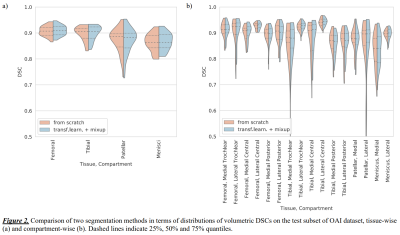
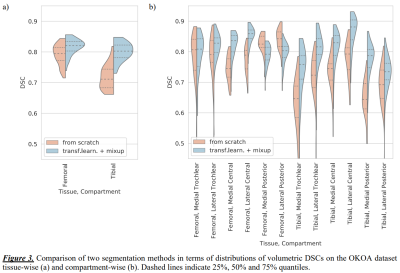
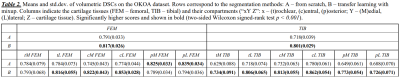
Tissue-wise volumetric DSCs showed that method B significantly outperformed A on both datasets (Table 1 and 2, Figure 2a). On OKOA both methods yielded lower relative segmentation accuracy due to the differences in MRI data acquisition protocols 3, yet method B showed better robustness (Figure 3a).We visually inspected the compartmentalization results and confirmed that they were consistent and in agreement with the multi-atlas.
Compartment-wise, both methods performed similarly on OAI (Table 1, Figure 2b), yet method B significantly improved (p<0.05) the DSCs for 4 compartments. On OKOA, method B achieved significantly higher (p<0.001) DSCs in most of the compartments (Table 2, Figure 3b). Method A was significantly more accurate in the posterior areas of femoral cartilage.
Discussion and Conclusions
In this work we proposed a new multi-stage method for compartment-specific segmentation of knee cartilage tissues from MRI data. Using this method, we were able to perform fine-grained comparison of tissue-level segmentation models and showed how such analysis can be used to select the most accurate model for the considered problem.Our segmentation approach showed performance superior to the previous state-of-the-art at both tissue and compartment levels. Notably, the improvements were observed in femur load-bearing areas on OAI and in most of the compartments on OKOA.
Even though the study considered only knee MRIs, it can be further extended to other domains. Particularly, the problems, where a trade-off between segmentation accuracies in multiple anatomical regions has to be made, would directly benefit from our approach.
Acknowledgements
The OAI is a public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2- 2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corpora- tion, GlaxoSmithKline; and Pfizer, Inc. Private sector fund- ing for the OAI is managed by the Foundation for the Na- tional Institutes of Health. This manuscript was prepared using an OAI public use data set and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
The authors would like to acknowledge the following funding sources: strategic funding of University of Oulu (Infotech Oulu), Sigrid Juselius foundation, and KAUTE foundation, Finland.
References
- Tack, A., Mukhopadhyay, A., & Zachow, S. (2018). Knee menisci segmentation using convolutional neural networks: data from the osteoarthritis initiative. Osteoarthritis and cartilage, 26(5), 680-688.
- Tack, A., & Zachow, S. (2019). Accurate Automated Volumetry of Cartilage of the Knee using Convolutional Neural Networks: Data from the Osteoarthritis Initiative. In IEEE 16th International Symposium on Biomedical Imaging, 40-43.
- Panfilov, E., Tiulpin, A., Klein, S., Nieminen, M. T., & Saarakkala, S. (2019). Improving Robustness of Deep Learning Based Knee MRI Segmentation: Mixup and Adversarial Domain Adaptation. In Proceedings of the IEEE International Conference on Computer Vision Workshops (pp. 0-0).
- Karani, N., Chaitanya, K., Baumgartner, C., & Konukoglu, E. (2018, September). A lifelong learning approach to brain mr segmentation across scanners and protocols. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 476-484). Springer, Cham.
- Peterfy, C. G., Schneider, E., & Nevitt, M. (2008). The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis and cartilage, 16(12), 1433-41.
- Data from 3D cartilage/meniscus segmentations of knee MRI scans. https://oai.epi-ucsf.org/datarelease/iMorphics.asp
- Podlipská, J., Guermazi, A., Lehenkari, P., Niinimäki, J., Roemer, F. W., Arokoski, J. P., … Saarakkala, S. (2016). Comparison of Diagnostic Performance of Semi-Quantitative Knee Ultrasound and Knee Radiography with MRI: Oulu Knee Osteoarthritis Study. Scientific reports, 6, 22365. doi:10.1038/srep22365
- Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
- Zhang, H., Cisse, M., Dauphin, Y. N., & Lopez-Paz, D. (2017). mixup: Beyond empirical risk minimization. arXiv preprint arXiv:1710.09412.
- Hunter, D. J., Guermazi, A., Lo, G. H., Grainger, A. J., Conaghan, P. G., Boudreau, R. M., & Roemer, F. W. (2011). Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthritis and cartilage, 19(8), 990–1002. doi:10.1016/j.joca.2011.05.004
- Klein, S., Staring, M., Murphy, K., Viergever, M. A., & Pluim, J. P. (2009). Elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging, 29(1), 196-205.
- Jones, E., Oliphant, T., & Peterson, P. (2001). SciPy: Open source scientific tools for Python.