2650
Deep-learning Methods for Meniscus Segmentation in Knee MRI1CUHK lab of AI in radiology (CLAIR), Department of imaging and interventional radiology , The Chinese university of Hong Kong, HongKong, Hong Kong
Synopsis
Meniscus segmentation in MR images has important clinical applications. Due to its large shape variation and low contrast with surrounding pixels, it is challenging to perform robust and reliable meniscus segmentation automatically. We investigated deep-learning network V-Net approaches for meniscus segmentation using MR images.
Introduction
The meniscus plays an important role in knee function. It not only reduces the load by absorbing the force but also reduces the friction and protects the articular cartilage during joint loading. Long-term stressed state makes meniscus prone to damage, especially for the athletes. Accurate segmentation of the meniscus has important clinical applications, including volume quantification and improved meniscal tear detection [1]. In recent years, deep-learning-based methods have made great progress and showed a significant potential in medical image segmentation. In this work, we investigated and compared 2D and 3D deep learning neural networks for meniscus segmentation.Methods
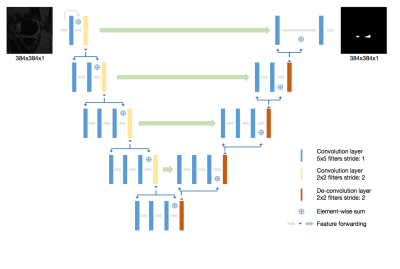
To the best of our knowledge, V-Net [2] has never been applied in meniscus segmentation using 3D MRI images. In this study, we performed both 2D V-Net and 3D V-Net [3] for meniscus segmentation and compared their performance. The knee MRI data sets including 88 subjects are from the Osteoarthritis Initiative (OAI) (https://oai.nih.gov). The MR images were acquired using a 3D double-echo steady-state (DESS) pulse sequence at 3T. The meniscus region was manually segmented by two expert radiologists. Further details of data can be found in OAI website. V-Net [2] architecture is a 3D deep learning neural network proposed for medical image segmentation. Different from another popular segmentation architecture U-Net, V-Net learns a residual function. The input of each stage in V-Net is used in the convolutional layers and added to the output of the last convolutional layer of that stage [2]. In order to extract better features, suitable for meniscus segmentation in 2D plane, a self-designed 2D V-Net model is proposed to segment meniscus. In our 2D V-Net model, the kernel size of convolutional layers and de-convolution layer are all reduced to two dimension compared to 3D V-NET. The network architecture is shown in Figure 1. Both models include a batch normalization layer between each convolutional layer and the activation function. Both the 2D and 3D V-Net were trained with a batch size of 5. The dice coefficient was used as the loss function to evaluate the similarity between output of network and the ground truth. Both 2D V-Net and 3D V-Net models used 100 epochs of training with initial learning rate of 1e-2. The learning rate decayed per 100 steps with the decay rate 0.99. The Adam optimizer was used during training process. The models were built using the open source Tensorflow platform and was running on a Titan V GPU card. Considering the resources of the computation, all 3D MR slices were patched to 256x256x32 as the input of 3D V-Net model. The segmentation result were the same dimension as input image after training process.Results
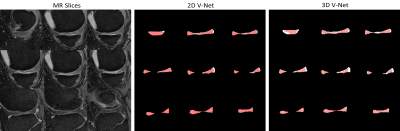
Figure 2 and Figure 3 show the results. The score of Sorensen dice coefficient for 2D V-Net and 3D V-Net model are 0.857 and 0.833 respectively. Figure 2 shows 2D plane of segmentation results slice by slice. Note the segmentation results from 2D V-Net have better resemblance with the ground truth and less false-positive pixels compared to 3D V-Net. Figure 3 shows the 3D volumetric meniscus segmentation results. Note 3D V-Net have smoother boundaries than 2D V-Net.Discussion and Conclusions
Accurate segmentation of meniscus is required for quantitative analysis of meniscus including volume, shape and position. In our study, we investigated deep-learning neural networks for automatic meniscus segmentation using MR images. Due to its large shape variation and low contrast with surrounding pixels, the manual intervention of radiologists are often needed when performing meniscus segmentation. As shown in Figure 2 and 3, the V-Net based models can effectively discriminate meniscus tissue from surrounding tissues. The results demonstrate that the 2D network can better extract 2D image features without being disturbed by spatial information. Meanwhile, the segmentation results of 3D model have better spatial smoothness by learning spatial information. Owing to discard of global information, 2D model could generate wrong disconnected regions in 3D visualization. Our study showed that the deep-learning-based network can achieve encouraging performance in automatic segmentation of meniscus. Notably, 2D network can improve performance in 2D plane but relinquish the spatial smoothness. Further work is needed to effectively combine the advantage of 2D and 3D deep-learning models.Acknowledgements
This study is supported by a grant from the Research Grants Council of the Hong Kong SAR (Project SEG CUHK02), and a grant from the Innovation and Technology Commission of the Hong Kong SAR (Project MRP/001/18X).References
[1] Fripp, Jurgen & Bourgeat, Pierrick & Engstrom, Craig & Ourselin, Sébastien & Crozier, Stuart & Salvado, Olivier. (2009). Automated Segmentation of the Menisci from MR Images. Proceedings - 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI 2009. 510-513. 10.1109/ISBI.2009.5193096.
[2] Milletari, Fausto, Nassir Navab, and Seyed-Ahmad Ahmadi. “V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation.” In 2016 Fourth International Conference on 3D Vision (3DV), 565–71, 2016. https://doi.org/10.1109/3DV.2016.79.
[3] Ko, Jacky KL. Implementation of Vnet in Tensorflow for Medical Image Segmentation. Github, 2018. https://github.com/jackyko1991/vnet-tensorflow.
Figures