2648
Kidney segmentation using dual neural network and one-shot deep learning1Radiology, UT southwestern medical center, Dallas, TX, United States
Synopsis
Image segmentation is important for quantitative analysis in many medical applications. Although deep learning methods generate segmentation with high accuracy, the model has to be trained for different organs and image modalities. These training processes are very time-consuming and labor-intensive because of the requirements of drawing masks manually. In this project, we present a dual neural network method trained using one set of 3D MRI data from a single subject. We demonstrate the feasibility of using a single 3D dataset to train a dual neural network for kidney segmentation, which greatly reduce the burden of drawing masks for large datasets.
Introduction
Deep convolution neural network (DCNN) is a powerful tool when trained using massive amounts of data. Image segmentation is crucial to quantification in many medical applications. DCNNs for segmentation have to be trained for different organs and image modalities1,2. However, DCNN training requires a large number of manually segmented masks, which is very time-consuming and labor-intensive, and may take considerable expertise. In most instances, there are very few segmented clinical image datasets available for training. The shortage of labeled data is the major challenge in wide clinical applications of DCNN-based segmentation methods. Different strategies have been employed to address this challenge using one or a few datasets 1,3. In this project, we investigated the feasibility of training a dual neural network to achieve acurate kidney segmentation using a single set of labeled 3D MRI data.Methods
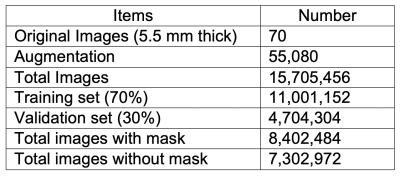
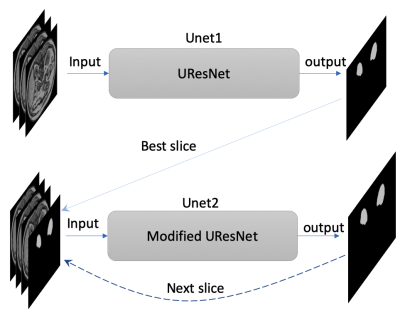
The labeled datasets were downloaded from the Combined Healthy Abdominal Organ Segmentation (CHAOS) challenge4. The MRI 3D dataset of in- and out-phase T1-weighted (T1w) images (a total of 70 images) from the first subject were used for training and validation. Since this dataset has a limited number of images, data augmentation plays a very important role in training. We took advantage of 3D properties of MRI data to perform 3D-rotational augmentations with 765 different rotations, besides regular 2D augmentations including radial distortion (x2), shear (x3), different noise addition (x6), and intensity inversion (x2). Total number of combinations for all these augmentations is 55080 (Table 1). The original T1w images with slice thickness of 5.5 mm were first resampled to isotropic images with a 1.89 x 1.89 x 1.89 mm3 resolution. These isotropic images were further augmented using different combinations of the aforementioned transformations. A dual neural network includes one UResNet (Unet1)5 and one modified UResNet (Unet2) to improve the robustness of mask prediction in Figure 1. Three slices with a slice gap of 5.67 mm were used as inputs for both Unets. In addition, the inputs of Unet2 also include a mask for the third slice. During the prediction phase, the mask input in the Unet2 was first generated by the Unet1 for the best predicted slice, and the rest input masks were generated by the Unet2 itself in Figure 1.The two Unets were trained separately using the augmented data from the first subject. The hyperparameters include: epoch=10, batchsize=50. Both Unets loaded the pre-trained imagenet weights as the initial weights. Dice coefficient was calculated as the training metric. 70% and 30% of the images were used for training and validation, respectively. In the prediction of the test stage, the masks for the first four subjects in CHAOS datasets were predicted by using the Unet1 and the dual Unets with the best trained weights. Total Dice coefficient for each 3D data set, slice-based Dice coefficients and the masked area for each slice were computed to evaluate the accuracy of the prediction.
Results
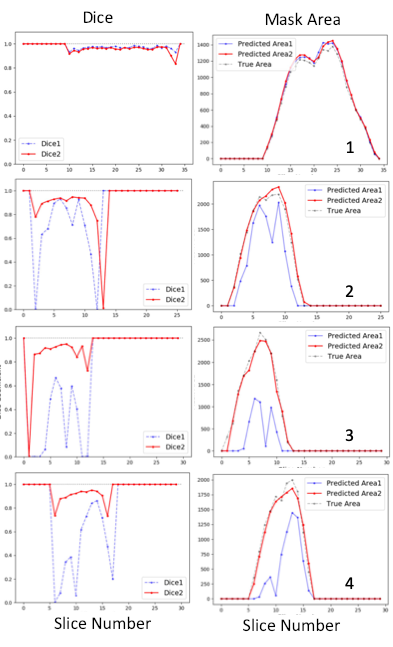
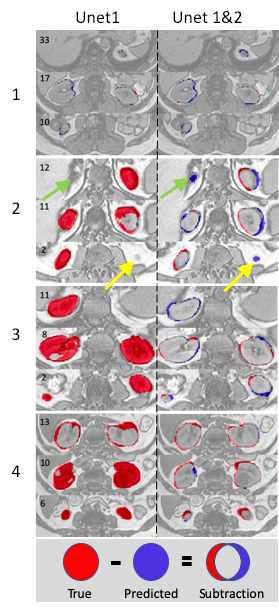
Figure 2. shows that both models generated nearly perfect masks for subject 1 (the trained subject) with total Dice coefficients of 0.972 and 0.96, respectively. For the rest three subjects, the dual Unets performed much better than the Unet1 only. For subject 2, total Dice coefficients were 0.915 for the dual Unets, vs. 0.744 for the Unet1 only; for subject 3, total Dice coefficients were 0.905 vs. 0.388; for subject 4, total Dice coefficients were 0.914 vs. 0.592. The numbers of the best predicted slices using Unet1 were 22nd, 9th, 6th, and 13th, respectively for the four subjects. The masks of these slices were used as the inputs for the Unet2. Thereafter, the Unet2 generated much better masks similar to the true mask in Figure 2. However, masks generated by the dual Unets may have larger inconsistencies (lower Dice values) with the true masks at upper and lower poles of kidney. Similarly, the slices at the upper and lower poles of the kidney usually represent the most difficult slices for manual drawing of the mask by humans.Figure 3 shows mask subtraction maps for the four subjects. One kidney or two kidneys may be missed by the Unet1 in the three new subjects, whose data the Unet1 didn’t see before in the training stage. In comparison, the dual Unets can segment both kidneys. The major differences compared to the true masks were at the edge of kidney. In subject 2, the dual Unets generated overestimated masks as pointed by the arrows, which may be missed by human due to partial volume effects (slice thickness of 9 mm for subject 2).
Discussion/Conclusion
Our results demonstrate the feasibility to train a dual Unets using one 3D dataset from a single subject to perform kidney segmentation on MRI. The prediction using the Unet1 needs further improvements, which could further increase the accuracy of the dual Unets. A few factors including image contrast and slice thickness may play very important roles in prediction of mask for other subjects. In conclusion, we achieved accurate prediction of kidney masks based on training using data from a single subject. This training strategy may be used for other clinical applications.Acknowledgements
This project is supported by NIH grants R01CA154475, U01CA207091, and P50CA196516.References
1. Zhang A., et al. Data augmentation using learned transformations for one-shot medical image segmentation. CVPR, June, 2019. https://arxiv.org/pdf/1902.09383.pdf
2. Wang K., et al. Automated CT and MRI Liver Segmentation and Biometry Using a Generalized Convolutional Neural Network. Radiology, 2019; https://doi.org/10.1148/ryai.2019180022
3. Zhang C., et al. CANet: Class-Agnostic Segmentation Networks with Iterative Refinement and Attentive Few-Shot Learning. CVPR, June, 2019. https://arxiv.org/abs/1903.02351
4. A CHAOS challenge. https://chaos.grand-challenge.org/Data/
5. Yakubovskiy P. Segmentation Models. https://github.com/qubvel/segmentation_models
Figures