2636
Travelling kidneys: Multicentre multivendor variability of renal BOLD and T1 mapping – preliminary results1Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, United Kingdom, 2Developmental Imaging and Biophysics Section, University College London, London, United Kingdom, 3Cambridge University Hospitals NHS Foundation Trust, Cambridge, United Kingdom, 4Imaging Biomarkers Group, Department of Biomedical Imaging Sciences, University of Leeds, Leeds, United Kingdom, 5Kidney Research UK, Peterborough, United Kingdom, 6Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, United Kingdom, 7Queen Square Institute of Neurology, University College London, London, United Kingdom
Synopsis
We have performed an intra-individual, inter-vendor comparison of renal T1 and BOLD T2* measures using predominantly product sequences. Renal T2* is sensitive to dephasing due to background magnetic field gradients, the degree of which differs across the MR vendors. A strong B0 L-R gradient, especially prominent for one vendor, leads to an asymmetry in T2* values between kidneys. B1 maps vary across subjects with no vendor-specific bias. Renal cortex T1 values computed using a 2-parameter fit were shown to systematically vary across vendors; a 3-parameter fit reduced this bias but differences remain. This highlights areas of future development for standardisation.
INTRODUCTION
Renal MRI has undergone significant developments in recent years, but standardisation and multicentre evaluation of measures is now crucial for clinical translation [1]. The UK Renal Imaging Network - MRI Acquisition and Processing Standardisation (UKRIN-MAPS) framework [2] aims to develop harmonized renal MRI protocols across MR vendors. This abstract presents initial results for measures of renal T1 and BOLD T2*, and characterization of scanner variability from B0 and B1 field maps.METHODS
Data Acquisition: Data was collected on four healthy subjects (age 33±8 years) at three UK sites each with a 3T scanner from a different MR vendor (GE, Siemens, Philips). Median time between scans of the same subject in each vendor was 24 days. The data was acquired using predominantly product sequences with near-identical acquisition parameters across vendors. All relaxometry data was collected with a 384x384x5mm2 FOV and 3x3x5mm3 voxel resolution.B0 field map: Dual-echo gradient echo scheme (echo shift ΔTE 3ms) acquired over the whole kidney volume in a single breath hold. B0 maps were collected with ‘default’ (over whole image) and ‘volume’ (over kidneys) shim settings using both a 2D and 3D readout scheme.
BOLD T2* mapping: Multiple gradient echo sequence acquired using in-phase echoes (TE1/ΔTE = 4.21/4.21ms, 12 echoes, 5 slices, fat saturation, 3 breath holds). Datasets were collected for both ‘default’ and ‘volume’ shim settings.
B1 field map: Acquired using the available B1 mapping scheme for each vendor (DREAM/ TurboFLASH B1 mapping /Bloch-Siegert method).
T1 mapping: Spin-echo echo-planar-imaging (SE-EPI) inversion recovery (IR) data acquired (single slice, SENSE2, FA90,TR 6000ms,TE 27ms) respiratory triggered with data collected at 10 inversion times (100/300/500/700/900/1100/1300/1500/2000/2500ms).
Data Analysis: B0 field maps were generated online (Philips, GE) or using FSL (Siemens). B1 field maps were generated using online software. BOLD T2* maps were fit to a log-linear decay function (for M0, T2*). Inversion recovery data was fit to a 2-parameter (for M0 and T1, assuming a perfect inversion) and 3-parameter fit (fitting for inversion efficiency, M0 and T1). Whole kidney ROIs were segmented manually from the magnitude data for both B0 and B1 maps, whilst cortex ROIs were generated for the T1 and T2* datasets. For each measure, the mode of the histogram of parameter values for each kidney was computed, from which the difference in measures between right and left kidney was calculated.
RESULTS
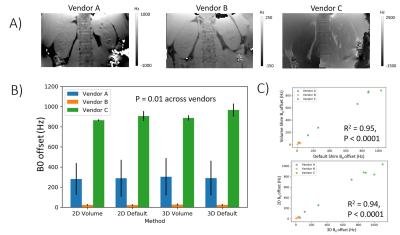
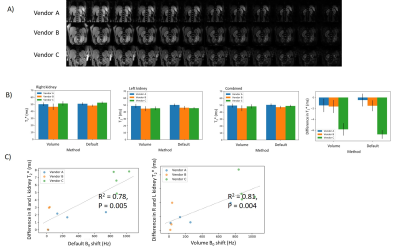
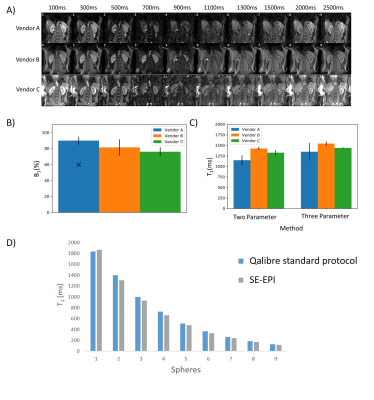
Figure 1 shows the L-R bias in B0 across the abdomen for each shim setting and vendor. There was a significant difference in the B0 L-R offset between vendors, with Vendor B consistently having the lowest offset and Vendor C the highest (Fig. 1B). There was a significant correlation of B0 L-R offset for ‘default’ and ‘volume’ shim settings, and for 2D and 3D B0 readout schemes (Fig. 1C). Figure 2 shows the BOLD T2* data, a clear systematic difference in T2* values is evident between left and right kidneys, with the greatest difference for Vendor C (Fig 2B). Figure 2C shows there was a significant correlation of the measured B0 L-R offset with T2* difference between left and right kidneys (default shim: R2 = 0.78, p = 0.005, volume shim: R2 = 0.81, p = 0.004), highlighting that the asymmetry in B0 results in T2* asymmetry. Figure 3A shows example T1 mapping data across inversion times, whilst Figure 3B shows the variation in the mean B1 (as percentage of requested flip angle) within the kidneys, this was not significantly different between vendors. T1 values were found to be consistently lower for Vendor A and C than Vendor B for the 2-parameter fit, as expected the 3 parameter increases the T1 values but discrepancy remains between vendors (Fig. 3C). Figure 3D shows the calibration of Vendor C against a standard of the Qalibre phantom.DISCUSSION
Here we assess the intra-individual, inter-vendor comparisons of renal T1 and BOLD T2* measures using predominantly product sequences. We demonstrate that renal T2* is sensitive to dephasing due to concomitant gradient magnetic fields, the degree of which is shown to vary across MR vendors. This B0 L-R gradient leads to an asymmetry in T2* values between L and R kidneys. Future work will address minimizing these B0 variations. B1 maps are shown to vary across subjects and vendors, but there was no vendor-specific bias. Renal cortex T1 values computed using a simple 2-parameter fit (assuming 100% inversion efficiency) were shown to systematically vary across vendors, as expected a 3-parameter fit reduced this bias however differences remain. It should be noted that this difference in T1 values may be due to the slice-selective inversion profile used by Vendors A and C, whilst Vendor B uses a non-selective inversion and is thus less prone to inflow effects, slice profile effects of the slice selective inversion and slice motion due to respiration.CONCLUSION
To understand the difference in inter-vendor relaxometry measures, characterisation of the B0 and B1 field maps is essential. Underlying B0 bias leads to alterations in renal T2*, which should be accounted for in analysis, particularly for multi-centre studies. Further optimisation of T1 measures are required to overcome systematic differences across vendorsAcknowledgements
This work is funded by MRC Partnership grant MR/R02264X/1.References
[1] Mendichovszky, I. et al. Technical recommendations for clinical translation of renal MRI: a consensus project of the Cooperation in Science and Technology Action PARENCHIMA. Magn. Reson. Mater. Physics, Biol. Med. (2019). doi:10.1007/s10334-019-00784-w
[2] Francis, S. et. al. UK Renal Imaging Network (UKRIN): MRI Acquisition and Processing Standardisation (MAPS). (2018). Available at: https://www.nottingham.ac.uk/research/groups/spmic/research/uk-renal-imaging-network/ukrin-maps.aspx. (Accessed: 10th July 2019)
Figures