2626
The effect of Compressed SENSE acceleration factor on 3D mDIXON sequence for liver imaging
Shiyu Wang1, Jiazheng Wang2, Qingwei Song1, Zhongping Zhang3, Bingbing Gao1, Lihua Chen1, and Ailian Liu4
1The First Affiliated Hospital of Dalian Medical University, Dalian, China, 2Philips Healthcare, Dalian, China, 3Philips Healthcare, Beijing, China, 4The First Affiliated Hospital of Dalian Medical University, Dlian, China
1The First Affiliated Hospital of Dalian Medical University, Dalian, China, 2Philips Healthcare, Dalian, China, 3Philips Healthcare, Beijing, China, 4The First Affiliated Hospital of Dalian Medical University, Dlian, China
Synopsis
Image acquisition of an mDixon sequence typically requires a breath hold to minimize respiratory motion artifact. Minimization the acquisition time of mDixon could be very useful in clinical daily routine. The purpose of the study is to explore the effect of the acceleration factor on the image quality of 3D mDIXON sequence for liver imaging using Compressed SENSE . And CS acceleration factor 4 can reduce the scanning time to 10s on the premise of ensuring the image quality, which is the best scheme for clinical 3D mDixon scanning of liver.
Introduction
mDixon technique exploits differences in chemical shift to separate the MRI signals from water and fat. Image acquisition of an mDixon sequence typically requires a breath hold to minimize respiratory motion artifact. However, breath-holding capacity can be limited and unreliable in children and patients of any age with dyspnea or tachypnea. Minimization the acquisition time of mDixon could be very useful in clinical daily routine. Compression sensing (CS) technology can obtain MRI image by optimizing signal processing algorithm in the case of k-space undersampling [1]. Compared with conventional methods, it can shorten the time of signal acquisition and become a research hotspot in the field of MR technology in recent years [2]. The purpose of this study is to investigate the effect of different acceleration factors of compressed sensing technology on the quality of 3D mDIXON sequence image of liver.Methods
Ten healthy volunteers(4 males, 25±4years) were recruited for liver scanning on a 3T whole‐body human scanner (Ingenia CX, Philips Healthcare, Best, the Netherlands) with a 16‐channel abdominal coil. Six groups of scanning schemes were designed: acceleration factor SENSE 2 and acceleration factor CS (2 / 3 / 4 / 5 / 6) were used for 3D mDIXON sequence scanning of the whole liver (the remaining scanning parameters were consistent, as shown in Table 1), and the scanning time was 20.5s, 18.7s, 12.9s, 10s, 8.5s and 7.1s respectively. Two observers placed ROI on the left outer lobe, left inner lobe, right anterior lobe, right posterior lobe at the level of the first hepatic hilum and the right vertical spinal muscle at the same phase direction at the same level, as shown in Figure one. The signal intensity and standard deviation of each lobe and vertical spinal muscle of the liver were measured respectively, and then the image SNR and CNR were calculated. Two observers scored the image quality of six groups on a 5-point subjective scale(table.2). The intra group correlation coefficient (ICC) test was performed on the data measured by two observers with SPSS 22.0. If the consistency is good, calculate the mean value of the measurement data and subjective scores of the two observers, and carry out subsequent statistical analysis. Kolmogorov Smirnov test was used to test the normality of the mean value of the data measured by the two physicians. According to the normal distribution data, one-way ANOVA was used, and LSD method was used to compare the following six groups. Kruskal Wallis H test was used to compare the non-normal distribution data among six groups and the following two groups.P < 0.05 was statistically significant.Results
The data and subjective scores of the two observers were in good agreement (ICC > 0.75)(Table.3). There was no significant difference in SNR and CNR between S2 and CS (2 / 3 / 4) sequences (P > 0.05). When the acceleration factor was greater than 4, the image SNR and CNR of CS5 / 6 group were significantly lower than that of CS (2 / 3 / 4) (P < 0.05); the image score of S2 and CS (2 / 3 / 4 /) sequence was better than that of CS (5 / 6) sequence (P < 0.05)(table.4,5).Discussion and Conclusion
Our study results showed that ten-second breath-hold CS-SENSE mDixon provided image quality comparable to that of the standard mDixon image. With the increase of acceleration factor, the scanning time is shortened while the SNR and image quality of liver are decreased. CS acceleration factor 4 can reduce the scanning time by about 50% on the premise of ensuring the image quality, which is the best scheme for clinical 3D mDixon scanning of liver.Acknowledgements
No acknowledgement found.References
[1] lusting m, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging [J]. Magn reson Med, 2007, 58 (6): 1182-1195. [2] geethanath s, Reddy R, Konar as, et al. Compressed sensing MRI: a review [J]. Crit Rev Biomed eng, 2013, 41 (3): 183-204.Figures
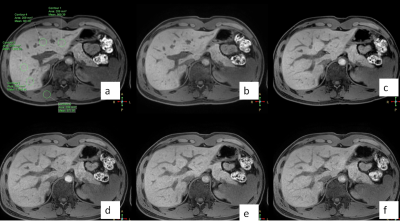
Figure one
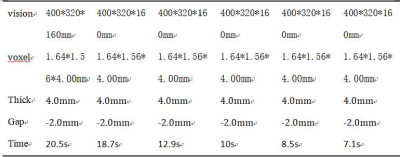
Scan parameters of each
sequence

Evaluation of image quality by five-point method

consistency test of six groups of quantitative data
and subjective scores of images by two observers (n = 10)
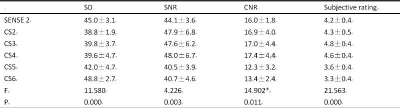
liver SD values, SNR and CNR and their comparison among the six groups (kruskal-wallis H)(n=10)
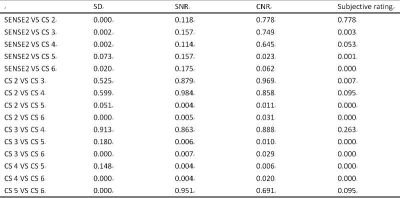
pairwise comparison results among the six groups (P value)