2601
Deep convolution neural network exploration for super resolution of abdominal 3D mDixon scans
Johannes M Peeters1 and Marcel Breeuwer1,2
1MR Clinical Science, Philips, Best, Netherlands, 2Biomedical Engineering – Medical Image Analysis, Eindhoven University of Technology, Eindhoven, Netherlands
1MR Clinical Science, Philips, Best, Netherlands, 2Biomedical Engineering – Medical Image Analysis, Eindhoven University of Technology, Eindhoven, Netherlands
Synopsis
Breath holding is often applied for abdominal imaging to avoid motion artifacts. However, breath holding limits the acquisition time and thus the resolution of the images. We propose to use 3D super resolution deep convolutional neural networks (CNN) to enhance the sharpness of 3D mDixon MRI. We found that sharpness increases with increasing number of network layers, but levels off already at 6 layers.
Introduction
Breathing motion is the main source of artifacts in abdominal imaging. Breath holding is commonly applied to minimize motion artifacts. It, however, limits the acquisition time window drastically and, therefore, a trade-off must be made with respect to coverage, resolution and SNR of the scan. Recently, super resolution based on convolutional neural networks (CNNs) has shown to be a promising technique to increase sharpness and thus reduce acquisition time. In this work, we investigate the use of CNNs to improve the resolution of breath hold 3D mDixon MRI at acceptable breath hold times.Methods
10 volunteers were scanned on clinical 3T MR systems (Philips Ingenia, Ingenia Elition). 3D mDixon MRI scans were performed, with breath holds times of 12.4 (scan 1) and 42 (scan 2) seconds, respectively. Both scans had a FOV of 450x400x249 mm and reconstructed voxels of 0.78x0.78x1.5 mm. Scan 1 had an acquisition voxel size of 1.8x1.8x3 mm (low resolution), scan 2 1x1x2 mm (high resolution). Compressed SENSE factor 5 was used to realize the breath hold times.The CNN training data consisted of the high-resolution scans 2 (training labels) and low-resolution scans derived from these scans (network input): all scans 2 were Fourier transformed, higher k-space frequencies were removed and ringing filtering was applied, followed by an inverse Fourier transform. This resulted in derived low-resolution acquisitions that match the sharpness of low-resolution scans 1.
3D data sets were split into 12420 patches of 32x32x32 pixels of which 9315 were used for training and validation (80/20%). Thereafter, testing was performed with the remaining 3105 patches outside the training and validation data. The CNNs tested were residual networks1 with 2, 4, 6, 8, 10 and 12 layers, each with 64 channels and 3x3x3 filter kernels, all trained with 200 epochs using the mean squared error (MSE) as loss function (Keras/Tensorflow, 2 NVIDIA 2080ti GPUs). Training time was 100 – 1000 minutes depending on the number of layers. Finally, all trained CNNs were applied to the short breath hold scans, i.e. the true low-resolution scan 1.
Results
In figure 1, the loss functions of all CNNs are shown. All show a gradual training loss decrease, but in case more layers are used, the validation loss increases after reaching a minimum, indicating overfitting. Network results for the test images (derived from the high-resolution reference) and differences with the high resolution reference of all CNNs are depicted in figure 2. All CNNs sharpen the images, which is clearly visible at the edges of the liver and the kidneys. The profiles in figure 3 indicate that as of 6 layers, the sharpness improvement decreases. An example of an axial and coronal reformat of a super-resolution image is shown in figure 4, using a truly acquired short breath hold image as input. These show that an improvement in all three voxel dimensions is realized, but also an increase in ringing artifacts.Discussion and Conclusion
Super resolution CNN can be a powerful solution to improve sharpness in scans with a limited acquisition time window like abdominal breath hold 3D mDixon MRI. A better delineation of organs like the liver and kidneys can thereby be achieved already with a limited number of layers in a residual network. It may allow to decrease the scan time such that more patients can comply with the required breath hold times. The residual artifacts and overfitting at a relatively low number of epochs indicate that the training dataset probably needs to be extended and that larger patches might need to be used. Also, this study was performed with volunteers who did not have any lesions. Next steps would be to check whether sharper lesion delineation can be realized with the trained networks as well as to check their robustness with contrast enhanced scans that are commonly applied with 3D mDixon for oncologic purposes.Acknowledgements
No acknowledgement found.References
1 Chaudhari et al., Super-resolution MSK MRI using deep learning, MRM 2018;80:2139-2154.Figures
Figure 1: Loss
functions of the trainings with the different number of layers
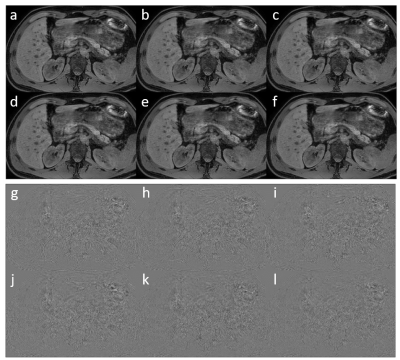
Figure 2: a-f
super resolution images of respectively 2, 4, 6, 8, 10 and 12 layer networks.
g-l respective difference with the high resolution image.
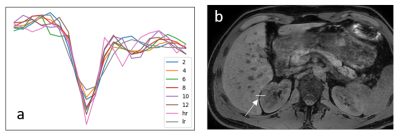
Figure 3: (a)
Profiles of the high resolution (hr), low resolution (lr) and super resolution images
(2-12) at the border between liver and kidney. The location of the profiles is shown
in (b).
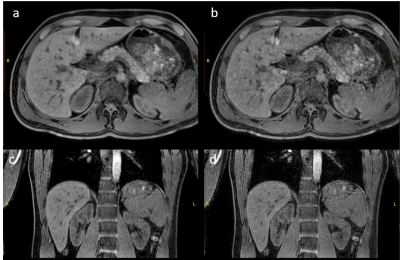
Figure 4: Axial
and coronal reformat of respectively the low resolution scan input (a,c) and 12
layer super resolution reconstruction (b,d).