2597
CT-based Synthetic pelvic T1-weighted MR image generation using a deep convolutional neural network (CNN)1Division of Radiotherapy and Imaging, The Institute of Cancer Research, London, United Kingdom, 2The Royal Marsden Hospital, London, United Kingdom, 3Department of Radiology, The Royal Marsden Hospital, London, United Kingdom
Synopsis
The MR-Linac exploits excellent soft-tissue contrast of MRI images with the option of daily re-planning for irradiation of pelvic tumours. However, this requires significant clinician interaction as contours need manual redefining for each radiotherapy fraction. Recently, machine learning-based segmentation approaches are being developed to automate this process. One major limitation of this approach is the lack of available fully-annotated MRI data for training. In this study, a deep learning model was developed and trained on CT/MR registered images of 21 patient datasets, generating synthetic T1-weighted MR images from input CT data, which could be useful towards disease segmentation on MRI.
Background
Conventional radiotherapy (RT) plans are performed using computed X-ray tomography (CT) scans for delineation of target volumes (PTVs) and organs at risk (OARs). These are subsequently used for optimal dose distribution, delivering maximal dose to the PTV and minimising irradiation of the OARs. The recent development of the MR-Linac provides a high-field magnetic resonance (MR) image-guided system for online RT treatment planning and adaptation, potentially improving the therapeutic ratio by reducing the spatial margins required to account for inter-fraction patient motion1,2. The excellent soft-tissue image contrast offered by MRI may also facilitate automated delineation of pelvic structures, including the target lesions and OARs during treatment of pelvic cancers. Machine learning-based approaches for automated segmentation, rely on adequately large training datasets. However, such datasets for pelvic MRI RT planning (images + ground truth delineations) are scarce. One solution for this may be to generate synthetic-MR (sMR) images from routinely-used CT scans, which commonly have associated delineations of relevant structures. Through this technique, previously annotated and segmented CT information can be transferred to sMR images which results in generation of larger training datasets, greater accuracy and elimination of manual delineation by radiologists for large ground-truth datasets.Purpose
To develop a convolutional neural network (CNN) model to synthesise pelvic T1-weighted MR images from CT dataMethods
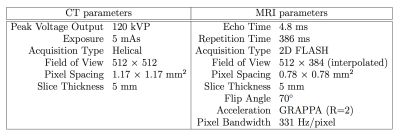
Patient Population and imaging protocolsInstitutional review board approval was obtained for this retrospective study. 37 datasets were identified from patients with lymphoma who underwent whole-body CT and MRI staging prior to conventional treatment. Scans were acquired a median of 6 days apart (range: 0-31 days) on a PET/CT scanner (Gemini, Philips) and 1.5T MR machine (Avanto, Siemens) respectively (Table 1). 16/37 datasets were excluded due to presence of metal hips or marked differences in patients positioning between CT and MRI, leaving 21 evaluable datasets (7 patients pre- and post-treatment and 7 patients pre-treatment only).
Spatial registration of MRI and CT scans
900 pelvic CT images were extracted from the datasets and registered to their visually selected corresponding T1-weighted MR image. To achieve accurate registration, the bed from CT images was removed using a patient-specific segmentation algorithm comprising of segmented CT bed pixel subtraction. Subsequently, the resulting CT images were aligned to the resampled MR images by centred transform initialisation and the paired images were then rigidly registered through 2D Euler transformation. Finally, the CT/MR images were non-rigidly registered using B-spline Free Form Deformation (FFD) (Mattes mutual information similarity metric: histogram bins = 15, Gradient descent line search optimiser parameters: learning rate = 5.0, number of iterations = 50, convergence minimum value = 1e-4 and convergence window size = 5) (Figure 1). 150/900 CT/MR images (3 full patient datasets and 33 further individual slices) exhibiting the worst CT/MR registration by visual inspection were reserved as a test dataset (excluded from the model training data), leaving 750 pairs of well-registered CT/MR images in the model training and validation data.
Model training
A U-Net architecture3 was trained to generate sMR images from input CT images. The image intensities, I, were normalised as follows:
CT: I’CT = (ICT + 1000)/2000
MR: I’MR = IMR / (IMR from the 97th percentile)
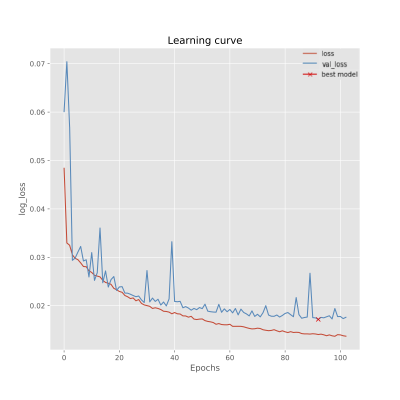
and then clipped such that no intensities fell outside the range (0, 1). The U-Net model was compiled after selection of an output layer consisting of a Relu activation function, using an Adam optimiser (learning rate = 0.001). To select an optimal loss function for the model, ‘Binary Cross Entropy’, ‘Mean Absolute Error (MAE) (L1)’ and ‘Mean Squared Error (MSE) (L2)’ were examined; L1 was selected as it yielded the lowest validation loss over 100 training epochs. The model was trained on a 1x Tesla GPU for 615 training images and 135 validation samples with batch size of 5. To prevent overfitting, early stopping was applied at the lowest validation loss and the best weights were selected (patience = 25).
Results
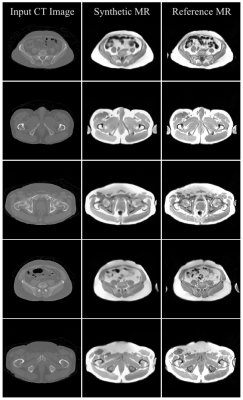
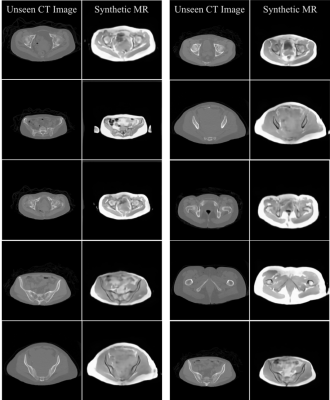
Preliminary results of the U-Net-based pelvic sMR generation model were promising. The model achieved minimum validation loss of L1=0.01717 (Figure 2). The results of our test dataset suggested that despite the relatively small dataset used, MR images could be regenerated from unseen CT images of new patients (Figures 3 and 4). Despite unsharpness in sMR images, the proposed model depicted promising correspondence and learning of soft-tissue features from the training data.Discussion and Conclusion
The preliminary regenerated MR images using the U-Net model in this study highlight the potential for improvement of the current model through the use of a larger dataset and advanced data augmentation. In addition, further optimising of hyperparameters and the CNN model architecture may yield to generating high-resolution pelvic sMR images. We conclude that synthetic pelvic T1-weighted MR images can be obtained from available patient CT data, which could be used to augment datasets for future segmentation models required for radiotherapy planning using the MR-Linac cancer treatment system.Acknowledgements
We acknowledge CRUK and EPSRC support to the Cancer Imaging Centre at ICR and RMH in association with MRC and Department of Health C1060/A10334, C1060/A16464 and NHS funding to the NIHR Biomedical Research Centre and the NIHR Royal Marsden Clinical Research Facility. This report is independent research funded by the National Institute for Health Research. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.References
1. Pathmanathan A, van As N, Kerkmeijer L, et al. Magnetic resonance imaging-guided adaptive radiation therapy: A “game changer” for prostate treatment?. Int. J. Radiat. Oncol. Biol. Phys. 2018; 100(2):361-373.
2. Burgos N, Guerreiro F, McClelland J, et al. Iterative framework for the joint segmentation and CT synthesis of MR images: application of MRI-only radiotherapy treatment planning. Phys. Med. Biol. 2017; 62:4237–4253.
3. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Miccai. 2015; 9351:234-241.
Figures