2539
Feasibility of Free Breathing Quantitative Susceptibility Mapping of the Liver: Comparison with a Breath-Hold Acquisition1Cornell University, Ithaca, NY, United States, 2Weill Cornell Medicine, New York, NY, United States, 3Northern Jiangsu People's Hospital, Yangzhou, China
Synopsis
Quantitative Susceptibility Mapping (QSM) enables accurate non-invasive monitoring of liver iron to guide iron-chelation therapy. Feasibility of a breath-hold sequence has been demonstrated in healthy subjects and in patients. In this work we investigate feasibility of a free-breathing navigator sequence in healthy volunteers to generate QSM maps and compare its correlation with the breath-hold sequence.
INTRODUCTION
Quantitative Susceptibility Mapping (QSM) is an emerging non-invasive MRI method to quantify iron, calcium and other susceptibility sources (1,2). In patients with transfusional iron overload, QSM enables accurate non-invasive monitoring of liver iron to guide iron-chelating therapy. Its feasibility using a breath-hold sequence (~ 30 sec) has been demonstrated in healthy subjects and in patients (3). However, in certain populations, such as pediatric and elderly patients, breath-hold compliance is lower leading to a higher failure rate. In this work, we investigate the use of a free-breathing navigator sequence to overcome the breath-hold requirement. In 6 healthy volunteers, we compared the QSM and R2* obtained using breath-hold and free-breathing data acquisitions.METHODS
Both breath-hold and free-breathing liver QSM acquisitions were performed on a 3 T scanner (Discovery MR750, GE Healthcare, Waukesha, WI) with a 32-channel body coil. In breath-hold (BH) sequence a multi-echo 3D gradient-recalled echo (GRE) sequence was used with the following imaging parameters: number of echoes = 5, unipolar readout gradients, flip angle = 3, TE1 = 2.9 msec, ΔTE = 3.4 msec, TR = 20.1 msec, reconstructed voxel size = 1.56 × 1.56 ×7 mm3, bandwidth (BW) = 390 Hz/pixel, acquisition matrix = 256 × 160 × (28-36), ASSET acceleration factor= 1.25, and acquisition time of 33–38 sec. The free-breathing (FB) sequence employed a pencil-beam diaphragmatic navigator to track respiratory motion (4,5). A 2-bin phase-ordered automatic window selection (PAWS) gating algorithm (4mm effective gating window) was used to tailor data acquisition according to diaphragm position in real-time (6). The remaining acquisition parameters were identical to the breath-hold acquisition with TE1=2.1ms, no acceleration and scan time of ~3 minutes.The T2*-IDEAL problem estimates fat content (F), water content (W), susceptibility induced field (f) and R2* decay by modeling the complex GRE signal shown in Eq. 1. We used out-of-phase echoes (∆TE= 3.4 msec at 3T) to calculate initial guesses (3).
$$(W,F,f,R_2^* )=argmin\sum_{j=1}^n|{S(t_{j})-e^{-R_2^*t_{j}}e^{-i2{\pi}ft_{j}}(W+F)e^{-i2{\pi}\nu_{f}t_{j}}}|^2_2 [1]$$
A QSM map was reconstructed using the MEDI algorithm (3,7),
$$\chi=argmin|{w(e^{-if}-e^{-i(d*\chi)})}|^2_2+{\lambda_1}|{M_{G}\triangledown\chi}|_1+{\lambda_2}|{M_{aorta}(\chi-\overline{\chi_{}}_{aorta})}|^2_2 [2]$$
assuming Gaussian noise (8), w the noise weighting, f the local field, d the dipole kernel, $$$M_G$$$ the binary edge mask, $$$\lambda_{1,2}$$$ regularization parameters, and $$$M_{aorta}$$$ the binary mask of abdominal aorta used for zero-referencing (9).
In all subjects 3 axial slices depicting approximately the same part of the liver were used for ROI analysis. 3 ROIs were drawn on the liver avoiding vasculature. R2*, and susceptibility values in the liver were calculated and compared between the BH and FB acquisitions. Linear regression (slope ,coefficient of determination ) and Bland-Altman analysis (bias and 95% limits of agreement LoA) were performed to compare the two methods.
Results
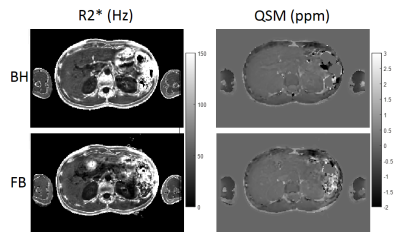
In all volunteers and both breath-hold and free breathing scans, data acquisition, water/fat separation and QSM reconstruction were performed successfully, with minor motion artifacts observed in one subject.In Figure 1, , and susceptibility maps in a healthy subject with both BH and FB scans show good agreement across the two scans
Figure 2 shows linear regression and Bland-Altman analysis in liver ROIs in R2* and QSM maps comparing breath-hold with free breathing acquisitions. R2* ranged from 35.7 to 76 Hz with good agreement between the two methods. QSM ranged from -0.23 to 0.19 ppm with good agreement among the two methods.
CONCLUSION
Our data indicate that free-breathing data acquisition is a viable option for patients who are unable to hold their breath during the scan.Acknowledgements
No acknowledgement found.References
1. Sharma SD, Hernando D, Horng DE, Reeder SB. Quantitative susceptibility mapping in the abdomen as an imaging biomarker of hepatic iron overload. Magn Reson Med 2015;74(3):673-683.
2. de Rochefort L, Liu T, Kressler B, et al. Quantitative susceptibility map reconstruction from MR phase data using bayesian regularization: validation and application to brain imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2010;63(1):194-206.
3. Jafari R, Sheth S, Spincemaille P, et al. Rapid automated liver quantitative susceptibility mapping. J Magn Reson Imaging 2019;50(3):725-732.
4. Nguyen TD, Spincemaille P, Weinsaft JW, et al. A fast navigator-gated 3D sequence for delayed enhancement MRI of the myocardium: comparison with breathhold 2D imaging. J Magn Reson Imaging 2008;27(4):802-808.
5. Nguyen TD, Spincemaille P, Cham MD, Weinsaft JW, Prince MR, Wang Y. Free-breathing 3-dimensional steady-state free precession coronary magnetic resonance angiography: comparison of four navigator gating techniques. Magn Reson Imaging 2009;27(6):807-814.
6. Jhooti P, Gatehouse PD, Keegan J, Bunce NH, Taylor AM, Firmin DN. Phase ordering with automatic window selection (PAWS): a novel motion-resistant technique for 3D coronary imaging. Magn Reson Med 2000;43(3):470-480.
7. Liu T, Liu J, de Rochefort L, et al. Morphology enabled dipole inversion (MEDI) from a single-angle acquisition: comparison with COSMOS in human brain imaging. Magn Reson Med 2011;66(3):777-783.
8. Liu T, Wisnieff C, Lou M, Chen W, Spincemaille P, Wang Y. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn Reson Med 2013;69(2):467-476.
9. Liu Z, Spincemaille P, Yao Y, Zhang Y, Wang Y. MEDI+0: Morphology enabled dipole inversion with automatic uniform cerebrospinal fluid zero reference for quantitative susceptibility mapping. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2018;79(5):2795-2803.