2482
Feasibility of Liver Fat Quantification based on the Threshold Extraction on fast 3D mDIXON images1The First Affiliated Hospital of Dalian Medical University, Dalian, China, 2Philips Healthcare, Beijing, China
Synopsis
The present study aims to explore feasibility of the threshold extraction method for liver fat quantification on the CS-SENSE 3D mDIXON-Quant images. Different from the conventional measurement of liver fat contents based on regions of interest that can be limited when the patients are associated with inhomogeneous fat distribution, liver fat quantification based on the 3D threshold-extraction can be more convenient and reliable.
Objective
To explore feasibility of the liver threshold extraction for liver fat quantification through the compressed SENSE 3D mDIXON-Quant.Introduction
Hepatic steatosis, usually the feature of fatty liver, is associated with pathological/abnormal accumulation of lipids in hepatocytes. Fatty liver (including alcoholic and non-alcoholic fatty liver) may also lead to chronic liver disease and cirrhosis. Therefore, measurement of fat fraction (FF) and assessment of hepatic steatosis will be of great value for clinical liver disease diagnosis and prognosis. Recently, the advanced compressed SENSE (CS) [1 ]MR 3D mDIXON-Quant sequence enables collection of whole liver fat map within one single breathhold. While the traditional post-processing method for measurement of whole liver fat fraction (based on regions of interest (ROIs)) can be complicated and time-consuming [2]. This study aims to explore the feasibility of a more convenient method for liver fat quantification based on the volume threshold extraction through images provided by the compressed SENSE 3D mDIXON-Quant.Materials and methods
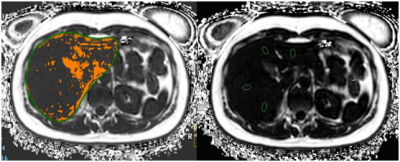
Study has been approved by the local IRB. 28 healthy volunteers (38.32±12.60) were recruited, and underwent the liver MR scans on a 3.0 T MR scanner (Ingenia CX, Philips Healthcare, the Netherlands) using the CS-SENSE 3D mDIXON-Quant sequence (CS-SENSE factor=2). On the vendor-provided post-processing workstation (IntelliSpace Portal, Philips healthcare), the threshold extraction and the traditional ROI-based methods were used by two observers for quantification of liver fat fraction for all volunteers. The threshold extraction method was carried out with the following steps: 1. the liver structure was automatically segmented on the 3D mDXION water-only image using the LIVER HEALTH software; 2. manually adjusted the segmentation, ensuring that all liver structure was included; 3. moved the segmentation to the fat-only image, and manually chose the threshold within a range of 0-1% to 5-8% (could be different among volunteers) to remove structures (shown in orange in Fig. 1) of blood vessels, bile duct, and surrounding fat tissue. For the traditional axial-ROI-based method, nine ROIs were positioned at reasonable slices of the liver, and the measured ROI-based mean values were taken into calculation of the liver fat fraction. The intra-class correlation coefficient (ICC) method was used to confirm the measurement stability between the two observers. The mean values of fat contents measured by the two observers were used to perform the Spearman test for evaluation of measurement correlation between the two methods.Results and Discussion
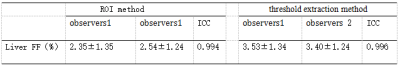
The fat fractions (Table 1) measured by the threshold extraction method from all the volunteers were (3.53±1.34) % and (3.40±1.24) %, respectively, by the two observers, while their counterparts for the traditional ROI-based method were (2.35±1.35) % and (2.31±1.13) %, respectively. The measurements show very good consistency between the two observers for each method (ICC=0.996, and ICC=0,994, P<0.05). A good correlation between the fat contents measured by the two methods are observed (r=0.997, P=0.001).And the significant difference between fat fractions measured by the two methods should be results from the 3D volume-threshold extraction and 2D ROI-based measurements. For the conventional ROI-based method, the liver fat content is represented by measurements from several specific regions, and it can be incorrect especially for those patients with inhomogeneous distribution of liver fat. Therefore, it is more feasible to use the 3D automatic threshold fitting method for entire liver fat measurements.Conclusion
The threshold extraction method shows fast and reliable determination of liver fat fraction for health volunteers based on the CS-SENSE 3D mDIXON-Quant images, and it may help to provide valuable information for the clinical screening, diagnosis and treatment-effect evaluation of liver diseases.Acknowledgements
No acknowledgement found.References
[1] Bratke G,Rau R,Weiss K,et al. Accelerated MRI of the lumbar spine using compressed sensing: quality and efficiency. J Magn Reson Imaging 2018; 49: 164-175.
[2] Vu KN, Gilbert G, Chalut M, et al. MRI-determined liver proton density fat fraction, with MRS validation: Comparison of regions of interest sampling methods in patients with type 2 diabetes. J Magn Reson Imaging, 2016, 43(5): 1090-1099.
Figures