2476
Development of a deep learning algorithm for detection of liver cancers on Magnetic Resonance Imaging
Sailong Zhang1, Keith Wan-Hang Chiu1, Siu Hin Mak2, TSOUGENIS Efstratios3, and Peng Cao1
1Diagnostic Radiology, The University of Hong Kong, Hong Kong, Hong Kong, 2The University of Hong Kong, Hong Kong, Hong Kong, 3Imsight Medical Technology Company (Hong Kong), Hong Kong, Hong Kong
1Diagnostic Radiology, The University of Hong Kong, Hong Kong, Hong Kong, 2The University of Hong Kong, Hong Kong, Hong Kong, 3Imsight Medical Technology Company (Hong Kong), Hong Kong, Hong Kong
Synopsis
Early detection of liver cancer is crucial for improving patient management outcome. However, liver lesions can be differencaited to be identified. In recent years, the fully convolution neural network (FCNN) has shown to be able to achieve commensurate and comparable performance of detecting various pathology on medical imaging. The goal of this study is to show the possibility of applying FCNN deep learning for training the hepatic lesion detection on dynamic contrast-enhanced Magnetic Resonance Imaging.
Purpose
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is the modality choice of detection and characterization of liver lesions1-3. Recently, with the advancement in deep learning, convolutional neural networks (CNNs) have shown to be capable of processing and analysis of medical imaging4. For the patient with liver disease, liver segmentation and tumor detection are very important for patient management. Although many machine learning approaches have generated compelling systems for the detection and staging of various lesions5, 6, the ability of deep learning on MRI or hepatic lesions still remains to be elucidated. Currently, the convolutional neural networks for liver lesion detection in radiology are mainly based on cropping the regions of interest (ROI) with boundary square box7, 8 , and some studies used bounding box for the lesion detection on T2-weighted MRI 9. Evolved from conventional CNN, FCN can increase the accuracy of feature detection by employing multi-resolution layers 10. V-Net11, as an improved version of U-Net12, is configured with 3D convolution that is capable of performing volumetric segmentation based on DCE liver cancer MRI. Here we propose a potential algorithm with V-Net on DCE-MRI for the hepatic lesion detection.Methods
Obtaining and labeling dataset100 abdomen MRI scans (by 3T Philips MRI with the same protocol) were retrieved from Picture Archiving and Communication System. Inclusion criteria include focal liver lesion and exclusion criteria include patients who have received local region therapy and MRI scans of non-diagnostic quality. Lesion contouring was accomplished with trained operators under the supervision of an experienced radiologist.
Deep learning
We apply V-Net11, a fully convolutional neural network (FCN) for volumetric segmentation. The training was performed using a GeForce RTX2080Ti graphic card. The network was implemented in Tensorflow (https://www.tensorflow.org/),. V-Net has 5 levels with 5 x 5 x 5 convolutional layers, 2 x 2 x 2 downsampling and de-convolutional layers. The feature dimension varied from 4 to 64 for levels 1 to 5. We rescale the image size from 576 x 576 x 149 (X, Y, Z) to 256 x 256 x 64 due to the hardware limitation. The whole training session was set up with a batch size of 5 and 200 epochs.
Model Validation
The neural network output was evaluated by three factors: lesion size, lesion location, and dice coefficient. The lesion size was measured by the longest diameter for each lesion contour.
Results and Discussion
With on-going progress, until now, 100 cases were successfully contoured and pooled into a training set and test set with a ratio of 8:2. A symbolic V-Net model with a maximum of 64 features has been built from 80 training cases. We set up a threshold of Cross-Entropy (CE) loss at 0.5 to control the effective output. In the model test, the validated prediction can reach up to 0.6 for CE loss, but with discrete spray pixels around most of the lesions (as Fig.3). The outcome is expected, for a starting training set at the initial stage, and with the incremental sample sizes, the FCNN can significantly improve the outcome with the 10th level (from n =80 to n = 800). This result suggests the possibility of utilizing FCNN like V-Net in deep learning training of DCE-MRI.Conclusion
Deep learning is suitable for medical scans which contain a huge amount of scan data, which ensures the sustainability of algorithm outcome, ruling out the bias such as over-fitting. Convolution neutral network can satisfy the 3-D processing for radiological tomography, and sustain the calculation with volume rendering. V-Net, with less time consuming, can attain outstanding features training outcomes for 3D images. Since the present study is still on-going, with more training sets, the system will incorporate more radiographic features. In summary, this on-going study at this stage introduces and elucidates a preliminary study to use FCNN algorithm for liver lesion detection with DCE-MRI.Acknowledgements
No acknowledgement found.References
- Albiin N. MRI of Focal Liver Lesions. Curr Med Imaging Rev. 2012;8(2):107-16.
- Burrel M, Llovet JM, Ayuso C, Iglesias C, Sala M, Miquel R, et al. MRI angiography is superior to helical CT for detection of HCC prior to liver transplantation: An explant correlation. Hepatology. 2003;38(4):1034-42.
- Lohrke J, Frenzel T, Endrikat J, Alves FC, Grist TM, Law M, et al. 25 Years of Contrast-Enhanced MRI: Developments, Current Challenges and Future Perspectives. Adv Ther. 2016;33(1):1-28.
- Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights into Imaging. 2018;9(4):611-29.
- Sato M, Morimoto K, Kajihara S, Tateishi R, Shiina S, Koike K, et al. Machine-learning Approach for the Development of a Novel Predictive Model for the Diagnosis of Hepatocellular Carcinoma. Scientific Reports. 2019;9(1):7704.
- Freiman M, Edrei Y, Sela Y, Shmidmayer Y, Gross E, Joskowicz L, et al., editors. Classification of Suspected Liver Metastases Using fMRI Images: A Machine Learning Approach. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008; 2008 2008//; Berlin, Heidelberg: Springer Berlin Heidelberg.
- Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29(7):3338-47.
- Wang CJ, Hamm CA, Savic LJ, Ferrante M, Schobert I, Schlachter T, et al. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol. 2019;29(7):3348-57.
- Esses SJ, Lu X, Zhao T, Shanbhogue K, Dane B, Bruno M, et al. Automated image quality evaluation of T2 -weighted liver MRI utilizing deep learning architecture. J Magn Reson Imaging. 2018;47(3):723-8.
- Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015.
- Milletari F, Navab N, Ahmadi S-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation2016. 565-71 p.
- Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; 2015: Springer.
- Davenport MS, Khalatbari S, Liu PSC, Maturen KE, Kaza RK, Wasnik AP, et al. Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology. 2014;272(1):132-42.
Figures
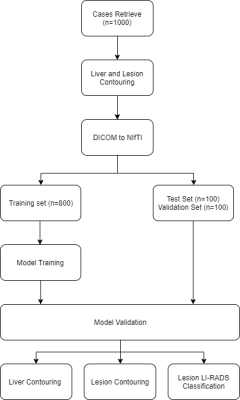
Flowchart of the model design in data preparation, training, and validation.
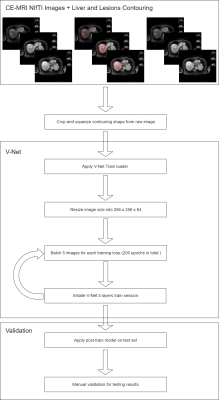
Infrastructure
of V-Net model.
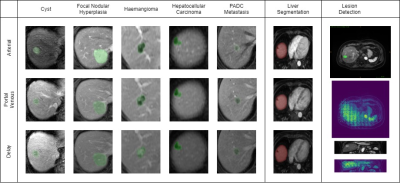
Sample contouring with various types of lesions in accordance with clinical radiology report, liver and lesion segmentation labeled with green and red color respectively. Samples of lesion detection have been attached in the final column.