2399
Evaluation of convolution strategies for 3D-CNN for MR Prostate segmentation1Department of Clinical Sciences, Intervention and Technology, Karolinska Institute, Stockholm, Sweden, 2Medical Radiation Physics and Nuclear Medicine, Karolinska University Hospital, Solna, Sweden, 3Imaging and Function, Karolinska University Hospital, Stockholm, Sweden, 4Breast Radiology, Karolinska University Hospital, Solna, Sweden, 5Department of Oncology-Pathology, Karolinska Institute, Stockholm, Sweden, 6Department of Molecular Medicine and Surgery, Karolinska Institute, Stockholm, Sweden, 7Department of Radiology, Capio S:t Göran Hospital, Stockholm, Sweden
Synopsis
Typical prostate MR dataset are 2D with few slices and high in-plane resolution. These dimensions do not conform naturally to traditional 3D convolution neural network up/down-sampling schemes, which customarily double/halves the input dimensions in all directions. We compared two strategies for up/down-sampling applied to prostate segmentation in MRI datasets: 1) Transform the datasets and applying orthodox convolution layers. 2) Adapt the up/down-sampling convolution layers in order to use more native resolution input datasets. Our results show that, in general, the former strategy works best, both in terms training loss convergence and overall segmentation results.
Introduction
Prostate segmentation on Magnetic Resonance Imaging (MRI) data is particularly difficult considering the organ’s non-distinct boundaries. In addition, axial T2-weighted MRIs of the prostate typically have large field-of-view for various other imaging considerations such as SNR and aliasing, hence 2D imaging with relatively few and thick slices are often acquired instead of 3D images with isotropic voxels.Convolution Neural Networks (CNN) have shown promise in medical image segmentation as they can be efficiently computed1 and require relatively few training data to produce useful results2. CNNs with UNET-based architecture typically halves/doubles the input data size in all dimensions at each down/up-sampling convolution layer3. This is not suitable for MR prostate data which typically have only few slices with relatively high in-plane resolution, resulting in limitations in how many convolution layers can be added to a network before data-reduction reaches a limit in the slice-encoding direction. One may then choose to either transform the input data such that all dimensions are of similar size to fit orthodox network architectures, or to adjust the up/down-sampling scheme of the network to accommodate native imaging dimensions.
Purpose
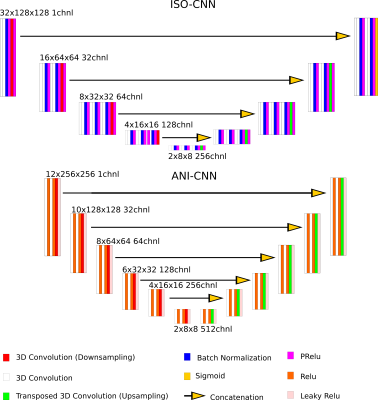
In this study, we compared the performance of two 3D CNNs for MR Prostate Images: One based on a traditional VNET encoder/decoder architecture when up-/down-sampling doubles/halves the input dimensions in all 3 directions (ISO-CNN), and another where the up-/down-sampling only doubles/halves the dimensions in the axial direction (ANI-CNN).Materials and Methods
Two different datasets were used for this study, one which is publicly available from the PROMISE12 challenge (https://promise12.grand-challenge.org/) and another self-acquired. Both datasets consist of expert manual segmentations on axial T2-weighted images. A variety of scanners, sequences, and acquisition parameters were used for acquisition. For our own dataset, 57 subjects were placed in training and 20 in the validation set. All 50 training examples in the PROMISE12 challenge were used as a part of the training dataset. This results in a training dataset size of 107 for the test set and 20 for the validation set.All datasets underwent common preprocessing pipelines for regularization and data augmentation (data augmentation for the training dataset only). The CNNs are implemented in Pytorch 10.1 and processed with CUDA 10.1 on a Nvidia GeForce 1080Ti GPU. Details of the CNNs are illustrated in Figure 1. Both CNN are trained using the same Adam optimizer and custom loss function defined as 1-DC where DC is the Sorensen-Dice Coefficient. The networks are trained with batch size 5, for 1200 epochs. The input data dimensions are 32x128x128 for ISO-CNN and 12x256x256 for ANI-CNN. Loss-progression, validation scores in the form of DC and The 95th percentile of the Hausdorff Distance (95th percentile of the set of minimum distances from one border to another) on the validation set, and visual inspection of predicted segmentations were used to evaluate the performance of the CNNs.
Results
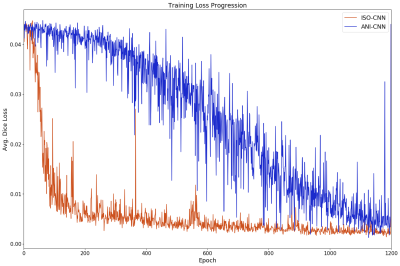
Figure 2 shows the training loss progression curves of the two networks. ISO-CNN “converges” much more quickly than ANI-CNN, with an overall faster and smoother descent with an average standard deviation of 0.047 for ISO-CNN and 0.13 for ANI-CNN.ANI-CNN achieved a Dice coefficient of 0.67 while ISO-CNN succeeded better with 0.81 on the validation dataset. The 95th percentile of the Hausdorff Distance [FS1] [YW2] for the validation dataset is 128.66 for the ANI-CNN and 68.57 for ISO-CNN.
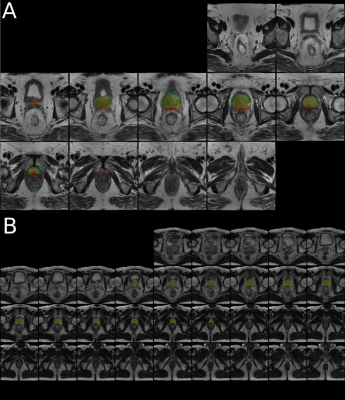
Figure 3 and 4 shows mosaic axial slices of a randomly selected examples from the validation dataset with automated segmentation from ANI-CNN (Fig. 3A) and ISO-CNN (Fig. 3B) together with the manual segmentation as ground truth. While ISO-CNN included of parts of the rectum at the apex of the prostate, it seems to generate a more correct prostate boundary. This issue is not present in all validation examples (Fig. 4).
Discussion
Results from this evaluation will act as a basis for future work when deciding which network will act as a base, and toward which direction, the optimization should occur for a more tailored CNN for prostate MR segmentations.Training was stopped prematurely after 1200 epochs for both networks due to time and resource constraints. Both networks’ loss progression still exhibits downward trend at Epoch 1200 (Fig. 2). This is especially true for ANI-CNN. The issue of parts of the rectum being included for ISO-CNN may be remedies by training the network further, but it may be also an inherent weakness attributed blurring due to oversampling in the slice-encoding direction.
A weakness of this comparison study is that no clear distinction can be made between CNN architecture or input size as cause of any differences between the two networks in terms of metrics and loss progression, but at the same time the purpose of the study is to evaluate networks which are tailored for their respective input data dimensions with focus not on the metrics, but rather visual inspection of the results.
Conclusion
We have evaluated two up-/down-sampling approaches to CNN-based deep-learning architectures applied to prostate segmentation on MR images. We have shown that orthodox up/down-sampling convolution layers of doubling/halving input data dimensions converge much smoother, and faster, than its anisotropic counterpart. It also performs better both in evaluation metrics and visual inspection.Acknowledgements
No acknowledgement found.References
1. LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998). Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11), 2278-2324.
2. Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
3. Kamnitsas, K., Ledig, C., Newcombe, V. F., Simpson, J. P., Kane, A. D., Menon, D. K., ... & Glocker, B. (2017). Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical image analysis, 36, 61-78.
Figures