2391
Applying Radiomics and Convolutional Neural Network Analysis to distinguish Lymph Node Invasion in Prostate Cancer using Multi-parametric MRI1Shanghai Key Laboratory of Magnetic Resonance, East China Normal Univeristy, Shanghai, China, 2Department of Radiology, the First Affiliated Hospital with Nanjing Medical University, Nanjing, China, 3Shanghai University of Medicine & Health Sciences, Shanghai, China, 4MR Scientific Marketing, Siemens Healthcare, Shanghai, China
Synopsis
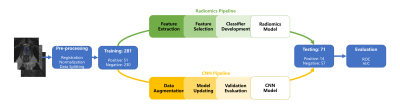
We compared a radiomics model and a convolutional neural network (CNN) model to distinguish lymph node invasion (LNI) in prostate cancer (PCa) using multi-parametric magnetic resonance images (mp-MRI). We trained the models in 281 patients and evaluated them in another 71 cases. The radiomics/CNN model produced an AUC 0.741/0.722, sensitivity 0.769/0.692, specificity 0.690/0.845 for the differentiation of LNI, which showed potential for the diagnosis of LNI in PCa.
Introduction
Pelvic lymph node invasion (LNI) is associated with increased risk of clinical recurrence and decreased long-term survival in patients with prostate cancer (PCa).1 Pre-treatment identification of PCa spread to the LNI is a critical indicator for patient counseling, clinical staging, and appropriate treatment selection and planning. Although the use of Briganti nomogram, MSKCC nomogram or other predictive models are recommended, task difficulties remain about how to integrate available risk elements to predicted LNI accurately.2 Recently, growing evidence indicates that machine learning (ML) methods could be a potential alternative to conventional methodologies with higher diagnostic effectiveness. Radiomics and convolutional neural networks (CNN) are two currently major methods to build feature based models for diagnosis.3,4 Therefore we built Radiomics and CNN model for LNI diagnosis, and compared their performance.Methods
We selected 281 cases (positive: negative =51: 230) as the training cohort. Another 71 cases (positive:negative = 14:57) were used for testing. We selected T2 weighted (T2W) images, diffusion weighted images (DWI) with b-value=1500 s/mm2, and apparent diffusion coefficient (ADC) images as the input data. All MR images were scanner on 3T MR scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany). First, we aligned the DWI and ADC onto the T2W images using Elastix.5 One radiologist drew the region of interest (ROI) of the PCa. Radical Prostatectomy together with pelvic lymph node dissection (PLND) or extend PLND was carried out in the form of open or laparoscopic approach.For the radiomics model, we used PyRadiomics (https://www.radiomics.io/pyradiomics.html) to extracted a total of 2553 features including shape, histogram, texture, and wavelets.6 Then we build a classification model as the following steps: a) Feature matrix was normalized submitting the meaning and dividing the standard deviation; b) Redundant features were reduced by a threshold value of 0.9 of Pearson correlation coefficient; c) The features were selected further by recursive feature elimination (RFE) with random forest (RF) algorithm; d) We selected the optimized model using one-standard rule on the 5-fold cross-validation. All above were implemented by FeAture Explorer (FAE, https://github.com/salan668/FAE), which was designed based on scikit-learn.
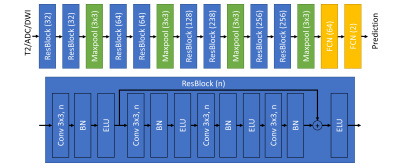
For the CNN model, we extracted a patch with shape of 160x160 containing the manual ROI. Patches of three sequences were stacked on the channel direction and combined to make a sample for model training. We referred ResNet model to design our CNN model. The architecture was shown in Figure 1.7 To monitor the process of the model training and avoid over-fitting, we split the training cohort into model-training data set (245, positive:negative=44:201) and model-validation data set (36 cases, positive:negative=7:29). The model-training data set was used for weights updating and model-validation data set was used for hyper-parameter updating. We used Adam with look-ahead as the optimizer a focal loss as the loss function.8 In order to overcome the limits of data set, also applied data augmentation including shifting, zooming, flipping and adding bias of received field during the training. All above were implemented using Keras (2.2.6) with Tensorflow (1.14) and the CNN model was trained on one NVIDIA Quadro GV100 card for 2 hours.
We used the area under the receiver operating characteristic (ROC) curve (AUC) to evaluate the radiomics model and the CNN model on the independent testing cohort.
Results
Radiomics model could achieve the AUC of 0.979 on the validation cohort and 0.741 on the testing cohort. For CNN model, The AUC achieved to 0.857 on validation cohort and 0.772 on the testing cohort. We showed the ROC curve in Figure 2. We also showed the confusion matrix in Table 1. The sensitivity of radiomics model was similar to the sensitivity of CNN model. But the specificity of CNN model was higher than the radiomics model.Discussion and Conclusion
ML model showed the potential power on the diagnosis of LNI in clinical diagnosis. Compared with the radiomics model, CNN model could achieve similar sensitivity but much higher specificity, which may be related to the data augmentation and data balance in the training process of CNN model. Beyond this work, more pre-process and post-process could be explored to improve the model performance, such as generative adversarial network to synthesize more data. In addition, adopting other sequence information, such as dynamic contrast-enhanced images, or combining radiomics and CNN model may improve the model accuracy, and will be consider in our future work.Acknowledgements
This project was supported by the National Key Research and Development Program of China (No. 2018YFC1602800), and the National Natural Science Foundation of China (61731009, 81771816).References
1. Briganti A, Larcher A, Abdollah F, et al. Updated nomogram predicting lymph node invasion in patients with prostate cancer undergoing extended pelvic lymph node dissection: the essential importance of percentage of positive cores [published online ahead of print 2011/11/15]. Eur Urol. 2012;61(3):480-487.
2. Mohler J, Bahnson RR, Boston B, et al. NCCN clinical practice guidelines in oncology: prostate cancer [published online ahead of print 2010/02/10]. J Natl Compr Canc Netw. 2010;8(2):162-200.
3. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2015;278(2):563-577.
4. Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis [published online ahead of print 2017/03/17]. Annu Rev Biomed Eng. 2017;19:221-248.
5. Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging. 2010;29(1):196.
6. Van Griethuysen JJ, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77(21):e104-e107.
7. He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. arXiv:151203385. 2015.
8. Zhang MR, Lucas J, Hinton G, Ba J. Lookahead Optimizer: k steps forward, 1 step back. arXiv:190708610. 2019.
Figures