2322
Inter-reader variability in Breast MRI Radiomics1Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Department of Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Wien, Austria
Synopsis
The purpose of this study was to investigate the effect of choice of radiologist on radiomics in breast MRI. Two cohorts of 100 patients each had breast lesions segmented by 4 independent radiologists, and these segmented images used to produce 101 radiomic features. over 90% of all features were found to have good or excellent agreement across readers, and a similar number of highly significant U-test were found for all readers. Differences between reader interpretation exist, however, they would not appear to effect the ability to distinguish between benign or malignant lesions.
Introduction
The majority of radiomics is conducted using a sub-section of an entire scan. The region of interest (ROI) is typically the tumor isolated from surrounding tissue and can be created either by drawing by hand, using a seed-point region growing algorithm, or automatically generated using deep-learning techniques. The final-say as to whether the ROI is appropriate/accurate, however, ultimately comes down to a radiologist’s interpretation [1]. The work presented here aims to investigate variability in radiomics via the differences a radiologist makes in the radiomic analysis of breast tissue lesions. We aim to look at both the statistical differences caused by the ROIs, and the effect on the ability to classify patients as either benign or malignant.Methods
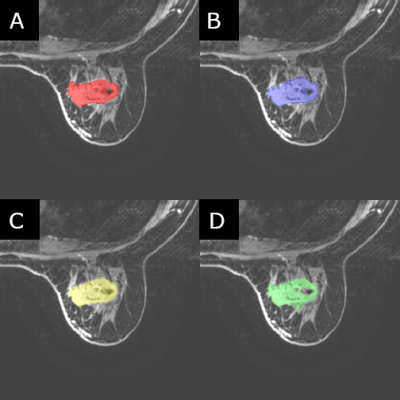
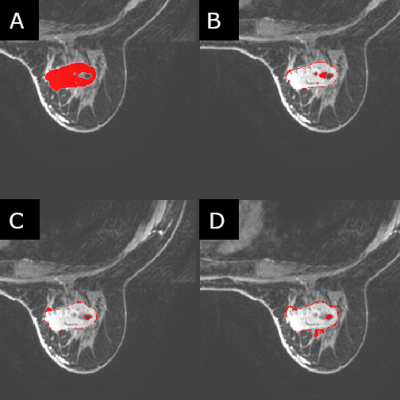
Two cohorts of 100 patients each were selected for this study where the two cohorts were defined as mass and non-mass lesions respectively. These were chosen to assess whether differences in radiomic parameters would be present in both “easy” cases (mass) as well as (or instead of) “hard” cases (non-mass). The patients in each cohort were classified as either benign or malignant based on the histology report following biopsy. Four trained radiologists segmented the identified lesions independently from each other, to the standard and accuracy which they deemed appropriate. Creating the ROIs on post-contrast contrast T1 images, all four readers used ITK-snap as their segmentation tool, with all four using a method of seed-growing/thresholding and/or manual segmentation for all 200 lesions. Radiomic data was calculated using CERR using a modified script in MatLab (2017b) to allow for batch processing of the images. 22 first-order and 79 higher-order statistics were calculated for each patient. Statistical analysis was conducted in SPSS (v.25) utilizing interclass correlation tests [2] to test for consistency between the readers, as well as ROC curves, and Mann-Whitney U-tests to test for significant parameters useful in classification.Results
When using the mass cohort, 87% (88/101) of radiomic features were found to have excellent agreement between readers (interclass correlation > 0.900) compared to 90% (91/101) in the non-mass cohort. These numbers increase to 91% (92/101) and 96% (97/101) respectively which were found to have at least good (ICC > 0.800) agreement. The number of significant (p<0.05), and highly significant (p<0.001), U-tests varied between readers, as did the number of significant (AUC>0.700) ROC curves. The difference between number of significant U-tests was greater for the non-mass cohort (range: 41 – 57) than in the mass cohort (range: 34-36), and the number of significant ROC curves also had a greater range (Range: 13 - 27) for the non-mass cohort when compared the mass cohort (Range: 24 – 28). The number of parameters which were highly significant for all readers varied between the mass and non-mass cohorts (25 and 13 features respectively), as too did the number of features which were found to be significant for at least one reader but not all four (9 and 26 respectively).Discussion
The high agreement across all test and high proportion of significant (and highly significant) univariate tests for all readers would suggest that the differences in feature extraction are negligible when taken as a whole. The aim of radiomics is to provide a discreet value to a patient so they can be measured against others, and to make classification possible, and a large variation in patient ranking would be evident by lower consistency across patients, parameters, and readers. A higher consistency across readers would be expected in the mass lesion cohort due to their simplicity, however, the higher number of parameters with excellent agreement, and highly significant U-tests, in the non-mass lesion cohort may be caused by the increased attention given to them by the radiologists, due to the intrinsic complexity of non-mass lesions to segment. In the setting of machine learning, where multiple parameters are used to classify patients automatically, techniques such as LASSO [3] or MRMR [4] are used to select the most significant parameters and increase performance of models without overfitting. With the number of highly significant parameters being greater than those which would be ideally used in a model for a cohort this size, feature selection would not be hindered in this regard, and so neither would model building. Classifying benign and malignant tumors in breast MRI is a relatively simple task for radiomics, and so the work presented here would need to be tested in other more challenging classification tasks to provide a more robust conclusion on the role of reader dependence.Conclusion
The choice of reader to segment tumors for radiomic studies will have an impact on the features that are calculated, however, the effect of these differences on the diagnostic ability to identify patients as benign or malignant when using a consistent reader is negligible.Acknowledgements
This work was funded in part by the Breast Cancer Research Foundation grant.References
1. Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1):36. Published 2018 Nov 14. doi:10.1186/s41747-018-0068-z
2. Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research [published correction appears in J Chiropr Med. 2017 Dec;16(4):346]. J Chiropr Med. 2016;15(2):155–163. doi:10.1016/j.jcm.2016.02.012
3. Fonti, V., & Belitser, E. (2017). Feature Selection using LASSO. Retrieved from https://beta.vu.nl/nl/Images/werkstukfonti_tcm235-836234.pdf
4. Radovic M, Ghalwash M, Filipovic N, Obradovic Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinformatics. 2017;18(1):9. Published 2017 Jan 3. doi:10.1186/s12859-016-1423-9
Figures
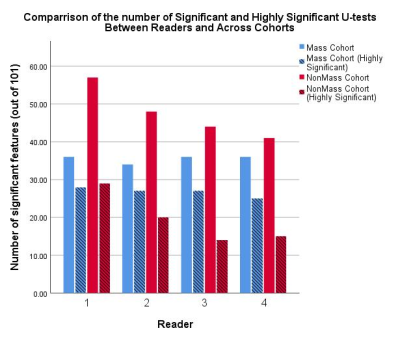
Figure 1: A comparison of the number of statistically significant (p<0.05, smooth bars) and highly significant (p<0.001, striped bars) Mann-Whitney U-tests for distinguishing between benign and malignant breast lesions. Variation between readers in the mass cohort is negligible, whereas variation between readers for the non-mass cohort are clear.