2313
Automatic Segmentation of Tumor-related Vessels of Breast Cancer on Ultrafast DCE MRI using U-Net1Radiology (Diagnostic Imaging and Nuclear Medicine), Kyoto Univ. Hospital, Kyoto, Japan, 2Faculty of Medicine, Kyoto University, Kyoto, Japan, 3Osaka Institute of Technology, Osaka, Japan, 4Kyoto University Graduate School of Medicine, Kyoto, Japan, 5Kyoto University, Kyoto, Japan
Synopsis
We aimed to develop a system for automatic segmentation of tumor-related vessels on ultrafast dynamic contrast (UF-DCE) MRI using U-net. Training set consisted of image dataset of 20 MIP images obtained from -15 to +60 sec after contrast injection from 59 patients. Exclusion criteria were those with poor image quality. The dice similarity coefficient was above 0.8 for training set, 0.6 for validation set. Careful analysis of failed cases revealed that inaccurate segmentation of the vessel were caused by low-contrast images, noisy-images, artifact, bright skin line due to incomplete fat suppression, and non-mass enhancement that may mimic vasculature.
INTRODUCTION
Ultrafast dynamic contrast enhanced (UF-DCE)-MRI of the breast captures very early phase of contrast enhancement of the lesion at a temporal resolution of 4 seconds. This state-of-the-art DCE technique allows us to evaluate vascularity of the lesion through up-slope kinetic information as well as vessels around the lesion, which are developed in case of aggressive malignant lesions [1]. The interval between feeding artery enhancement and drainage vein enhancement is associated with breast cancers. Breast cancer induces angiogenesis and this leads to tumor progression. Therefore, it is important to evaluate tumor-related vessels. Visual evaluation failed to capture quantifiable nature of the vessels. Manual segmentation are time-consuming and subjective, suggesting the need of quantification [2]. Deep convolutional neural network is one of the pixel-based machine learning methods. Among various network, U-Net is a fully convolutional network and has been used for image segmentation including breast fibrograndular tissues. In order to establish automatic system to quantify tumor-related vascularity, we aimed to develop U-Net system to automatically segment breast tumor-related vessels on UF-DCE MRI separate from the breast tumor itself.METHODS
The study population consisted of female patients who underwent breast MRI with UF-DCE-MRI from December 2015 to March 2018 due to known / suspected breast lesions. Images with poor image quality and without any enhancing lesion were excluded. For malignant lesions, In total, 46 patients who underwent UF-DCE MRI were included.MRI procotol: MR images were obtained with 3T MR unit (Prisma/Skyra, Siemens). Gadobutrol/Gadoteridol was intravenously infused (2.0ml/sec). UF-DCE MRI (15sec before -60sec after the contrast injection, 3.7sec×20frames) was followed by standard DCE-MRI. UF-DCE-MRI was acquired by a prototype based on the 3D gradient-echo VIBE sequence using a CS reconstruction (TR/TE 5.0/2.5, FA 15, FOV 360×360mm, matrix 384×269, thickness 2.5mm, CS acceleration=16.5), with 30 iterations. Based on these UF-DCE MRI, 19 subtracted MIP images per patient were computed.
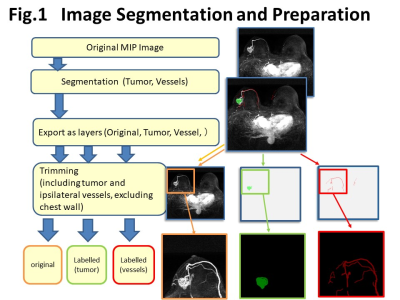
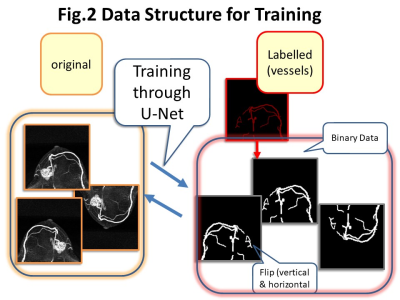
Image segmentation and training for U-Net: Out of 19 subtracted MIP images, the first 5-6 images were excluded from the analysis because no enhancing lesions were identified. The training data consisted of 364 MIP images from 32 patients for training and 174 MIP images from 14 patients for validation.The main lesion(s) and vessels were manually segmented by three readers (supervised by an expert breast radiologist). These segmented areas were exported as layers, then trimmed (fig.1). Images were trimmed to include tumor and ipsilateral vessels, and exclude chest wall. For current analysis, original images and labelled vessel images were used. The trimmed images were 128 x 128 pixel batch images per lesions. Image augmentation by horizontal and vertical flips were used for training set (fig.2). Deep learning segmentation was performed using the U-Net, consisting of convolution and max-pooling layer at the descending part, while convolution and up-sampling layer at the ascending part. The manually segmented images were used as a ground truth. The performance of segmentation was evaluated using the overall accuracy per pixel and the Dice Similarity Coefficient (DSC). Visual inspection of the segmented image created by U-Net were also performed.
RESULTS
Initial training using accuracy as evaluation function resulted in over 0.9 accuracy. However, actual vessel images were not ideal with over and under estimation of the segmented area. For more strict training, the further analysis used DSC as evaluation function, and “1-DCS” as loss function. Final model achieved a DSC of more than 0.8 for training set, yet DSC of validation set remained approximately 0.6.Cases of low DSC with “unsuccessful” segmentation were analyzed for possible reasons. Vessel-related reasons included small vessels that just started to enhance, small and tortuous vessels, vessels running over or close to the main lesion, the vessel adjacent to the non-mass type lesion. Non-vessel-related reasons included severe motion artifact, incomplete fat suppression, marked background parenchymal enhancement.
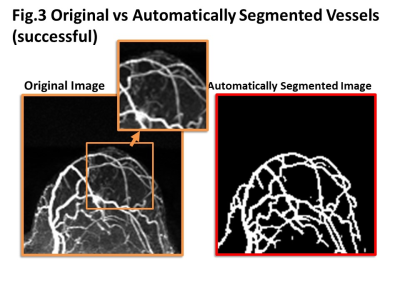
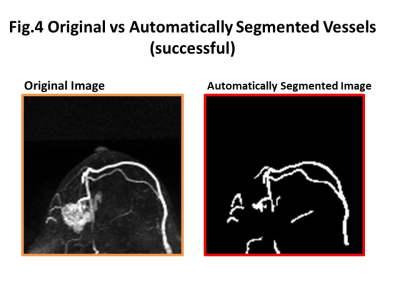
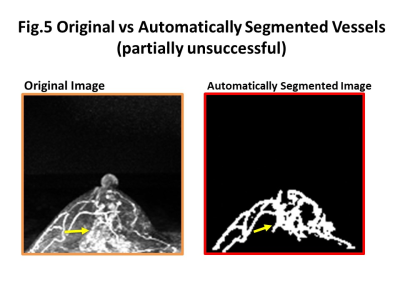
Some representative cases comparing original and automatically segmented images of the tumor and tumor-related vessels were shown in figure 3-5. In general, major thick vessels were successfully identified, while smaller thinner vessels were not detected by the trained automatic segmentation system.
DISCUSSION and CONCLUSION
This trial of automatic segmentation using U-Net demonstrated that automatic segmentation and quantification of tumor –related vessels is possible, yet challenging. The vessels in the breast are generally small. In addition, experimental results showed that tumor-related vessels are tortuous and incomplete, that makes segmentation harder. Further training to improve accuracy include increase the sample size, with more weight to identify thinner vessels.Acknowledgements
Dominik Nickel and Yuta Urushibata from Siemens for their technical support. KAKENHI-JP 15K09922References
1. Onishi, N., et al., Ultrafast dynamic contrast-enhanced mri of the breast using compressed sensing: breast cancer diagnosis based on separate visualization of breast arteries and veins. J Magn Reson Imaging, 2018. 47(1): p. 97-104
2. Wu, C., et al., Quantitative analysis of vascular properties derived from ultrafast DCE-MRI to discriminate malignant and benign breast tumors. Magn Reson Med, 2019. 81(3): p. 2147-2160.
Figures