2252
Stabilized decompositions for improved cardiac self-gating: A proof-of-concept in single breath-hold 3D cine imaging1Laboratory for Psychiatric Neuroscience and Psychotherapy, University of Fribourg, Fribourg, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3Advanced Clinical Imaging Technology, Siemens Healthcare, Lausanne, Switzerland, 4Center for Biomedical Imaging, Lausanne, Switzerland, 5LTS5, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Synopsis
ECG recording during whole heart MR imaging requires extra setup time and is not always reliable. Recently, self-gated (SG) acquisition techniques have been developed, which extract the cardiac signal from k-space center amplitude modulations. Here, we investigated whether different single- (Empirical Mode Decomposition (EMD), Ensemble EMD (EEMD)) and multi-coil (PCA, ICA with novel stabilisation) decomposition methods yielded SG triggers with minimal variability to ECG R-wave location. EEMD yielded median variability similar to PCA. Compared to PCA, our proposed stabilised ICA approach yielded SG triggers with 66% lower variability (median over 5 subjects), although not for all subjects.
INTRODUCTION:
Motion-resolved imaging1,2 of the heart depends on reliable physiological gating signals. ECG-triggering is commonly used for synchronising the MR acquisition to the cardiac cycle. Recently, self-gating (SG) methods have shown promise for simplifying the setup by instead extracting the cardiac motion information directly from the acquired imaging data3. Approaches for extracting self-gating signals have mainly rely on principal component analysis (PCA)1,2,4, or independent component analysis (ICA)1. However, PCA and ICA assume linearity and stationarity of the analysed time-series, which may not be realistic in cardiac imaging due to e.g. non-sinus rhythm, and ICA runs can yield very different components5. Therefore, we hypothesize that decomposition methods with less restrictive stationarity assumptions or more stability may result in improved cardiac SG signal extraction. Given varying coil distances to the myocardium, it is also uncertain whether all coils are needed to recover an SG signal, and therefore we explored both univariate methods (single-coil) and multivariate methods (multi-coils).METHODS:
Datasets:Single breath-hold (scan time=28s) 3D cine datasets were acquired in N=5 healthy volunteers on a 1.5T clinical MRI scanner (MAGNETOM Aera, Siemens Healthcare) using a free-running 3D golden angle radial bSSFP6,7 sequence. An ECG was simultaneously recorded for analysis purposes. The signals of interest were obtained by selecting the absolute value of the central coefficient of periodically acquired k-space readouts (K0 modulation).
Decomposition approaches:
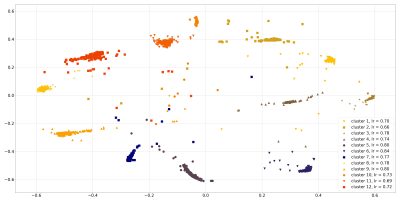
We tested single-coil and multi-coil decompositions. Single-coil decomposition were performed on all coils sequentially. Empirical mode decomposition (EMD), a single-coil method, is particularly suited for non-stationary signals, but can be affected by mode mixing. Thus, we also used ensemble EMD (EEMD), which improves over EMD by running the decomposition multiple times with added white noise, turning EMD into an adaptive dyadic filter bank. Averaging over the runs makes the method less sensitive to the perturbations to data8. For multi-coil decompositions, we used the ICASSO principle5 with FastICA9 implementation to obtain 12 components over 200 runs of ICA. We then clustered all components into 12 clusters (Figure 3) using hierarchical clustering with a Ward variance minimization algorithm.
We propose two automated approaches to picking a decomposition from the multiple ICA runs. In the centrotype ICA approach, we estimated the sources by averaging all components within each cluster. This centrotype was then used to compute “pseudo sources” from the original signals, with no independence guarantee. In the proposed second approach, stabilised ICA (StabICA), we selected the most reliable decomposition based on a cluster quality index Iq: the difference between average within-cluster(Cm) similarities σi,j and average between-cluster(C-m) similarities: $$ I_q(C_m) =\frac{1}{|C_m|^2}\sum_{i,j\in C_m} \sigma _{i,j} -\frac{1}{|C_m||C_{-m}|}\sum_{i\in C_m}\sum_{j\in C_{-m}} \sigma _{i,j}\text{; where }C_{-m} = C - C_m $$
These methods were compared to PCA as a baseline.
Automatic SG component selection:
For each dataset, the subject-specific heartrate was extracted using FFT as the frequency with maximum power in the 0.5-2Hz band. This frequency was later used to automatically identify the components of interest, by selecting the component with the highest power spectral density in the ±0.3Hz band around heart rate.
Evaluation metrics:
The timing differences between peaks of the SG signal and triggers obtained from ECG R-waves were computed for each of the around 25 heartbeats per dataset. The distance between SG peaks varies, due to delay between the electrical activity and heart motion, ECG filtering and sign indeterminacy of ICA. Thus, we used standard deviation of the distribution of timing differences as the evaluation measure. The distributions of standard deviations were compared subjectwise between methods using the robust Bonett-Seier test of scale for paired samples10.
RESULTS:
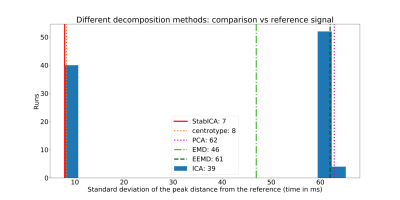
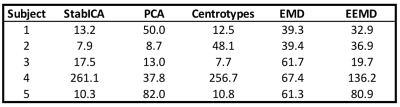
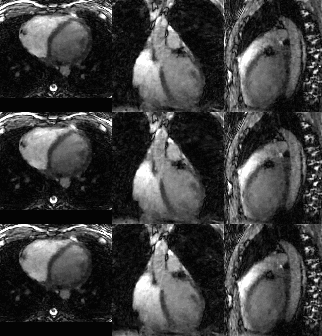
Most methods were able to recover a component representing the cardiac cycle. Specifically, StabICA, Centrotypes ICA, PCA, and EEMD recovered all triggers in 4 out of 5 subjects, while EMD recovered all triggers only in 2 cases. No method recovered all triggers for subject 4 (see Figure 3). The median standard deviation of the differences between SG triggers and ECG triggers across the 5 datasets were: StabICA: 13.2 ms, centrotypes: 12.5 ms, PCA: 37.8 ms, EMD: 61.3ms and EEMD: 36.9 ms. Figure 2 shows standard deviations for the various decomposition of one dataset. Two subjects had significantly lower variability with StabICA than PCA (p=0.005, p<10-16), one had lower variability but no significant difference (p=0.25), and two had higher variability with StabICA. Figure 3 shows results for all subjects. Qualitative examination of ECG, PCA, and StabICA reconstruction for one subject showed no visible quality difference (Figure 5).DISCUSSION/CONCLUSION:
Stabilized ICA and Centrotypes ICA showed lower variability than other decomposition methods. This supports our hypothesis that fewer assumptions might provide more “ECG-like” cardiac self-gating, as gating based on the ECG signal itself has no such assumption. However, the ECG as a reference standard is not ideal due to missed triggers or falsely identified triggers caused by MHD effects. More generally, our proposed approaches could improve over vector gating, which requires manual selection of the ECG vector and constant monitoring for missed triggers. Image quality on an example subject was not qualitatively different between SG decomposition methods, and future work includes expanding the analysis to more subjects including patients with arrhythmia, varying sequences, as well as solving sign indeterminacy of decompositions to further improve consistency.Acknowledgements
No acknowledgement found.References
1.Di Sopra L, et al. An automated approach to fully self‐gated free‐running cardiac and respiratory motion‐resolved 5D whole‐heart MRI. Magn. Reson. Med. 2019;82:2118–2132
2. Feng L, et al. 5D whole-heart sparse MRI. Magn. Reson. Med. 2018;79:826–838 doi: 10.1002/mrm.26745.
3. Larson A.C, et al. Self-Gated Cardiac Cine MRI Magnetic Resonance in Medicine 51:93–102 (2004)
4. Pang J, et al. ECG and navigator-free four-dimensional whole-heart coronary MRA for simultaneous visualization of cardiac anatomy and function. Magn. Reson. Med. 2014;72:1208–1217 doi: 10.1002/mrm.25450.
5. Himberg J, Hyvärinen A. Icasso: software for investigating the reliability of ICA estimates by clustering and visualization. In Proc. 2003 IEEE Workshop on Neural Networks for Signal Processing (NNSP2003), pp. 259-268, Toulouse, France, 2003
6. Piccini D, et al. Four-dimensional whole-heart cine imaging with isotropic resolution in one single breath-hold: initial results. Proc. ISMRM 2019.
7. Coppo S, et al. Free‐running 4D whole‐heart self‐navigated golden angle MRI: Initial results. MRM 2014
8. Zhaohua W, Norden E. Ensemble Empirical Mode Decomposition: a Noise-Assisted Data Analysis Method Advances in Adaptive Data Analysis, Vol. 1, No. 1 (2009)
9. Hyvärinen A, Oja E. Independent Component Analysis: Algorithms and Applications, Neural Networks, 13(4-5):411-430, 2000
10. Bonett D.G, Seier E. Statistical inference for a ratio of dispersions using paired samples. Journal of Educational and Behavioral Statistics, 28, 21-30, 2003
Figures
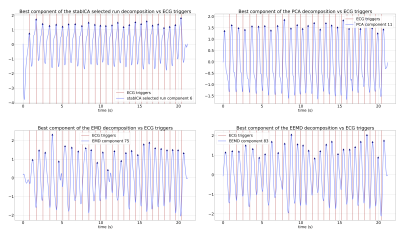
Example decompositions for one dataset (subject 1). For each of the decomposition methods, one component of interest was extracted, based on the previously established heart rate. The extracted component was then band-pass filtered to remove any high-frequency noise, yielding an SG signal estimate.
Component of interest for: (a) stabilised ICA (b) PCA (c) EMD (d) EEMD. Vertical lines mark ECG triggers (reference), blue lines show components with triangles showing the estimated SG triggers. EMD displays mode mixing (c), while EEMD largely eliminates it (d)