2238
Deep Learning Artifact Removal for Real-Time Rodent Cardiac MRI1Department of Internal Medicine II, University Ulm Medical Center, Ulm, Germany, 2Core Facility Small Animal Imaging (CF-SANI), Ulm University, Ulm, Germany
Synopsis
Reconstruction methods incorporating Deep Learning have gained a lot of traction in the recent past. However, most of these methods have been evaluated in human MRI. In this work, we show the feasibility of deep-learning artifact removal for the tiny golden angle radial trajectory in rodent cardiac MRI and validate two approaches against a Compressed Sensing reconstruction and the gated reference standard. The deep-learning based methods achieve acceptable visual image quality and exhibit only slight, but for one method significant, differences in the functional analysis.
Introduction
Cardiovascular magnetic resonance imaging has proven valuable for the assessment of structural and functional cardiac abnormalities. The long acquisition times of gated or self-gated techniques limit its widespread application in small animals. High undersampling has been applied and rising artifacts been addressed by compressed sensing (CS) or deep learning (DL) reconstructions1,2. Recently introduced convolutional neural networks (CNN) incorporate the temporal dimension into the learning process, which appears challenging in small animals, due to the limited number of cardiac phases reconstructable and high variability of the heart rates between 300 and 840 bpm3. We evaluate the application of tiny golden angle (tyGA)4 radial sparse MRI for real-time cardiac imaging in the mouse model with artifact removal based on a 2-dimensional convolutional neural network.Methods
All imaging experiments were performed on an 11.7 Tesla dedicated small animal MRI system (BioSpec 117/16, Bruker Biospin, Ettlingen, Germany), equipped with a four-element thorax coil (RAPID Biomedical, Rimpar, Germany). A stack of short axes orientations was used for volumetric evaluation. Reference data were acquired applying a conventional Cartesian self-gated sequence (IntraGate ©, ParaVision 6.01, Bruker), scan parameters are given in Table 1. Real-time imaging was based on the tyGA4 trajectory and reconstruction was performed off-line with an in-house built MATLAB framework in conjunction with Python and Tensorflow. Image series were either reconstructed by a k-t SPARSE-SENSE framework with a Total Variation sparsity operator or with a simple sliding-window approach and ensuing artifact removal by two different 2D CNNs. The structure of the networks is shown in Figure 1. Both networks were trained on synthetic data from previously acquired Intragate images in different subjects. The generation of artifact contaminated training data followed1. A total of 14960 training image pairs from 141 mice could be included. The network was trained for 350 epochs with the adaptive moment estimation algorithm and a validation split of 0.2. The study comprised eight male wild-type mice (C57BL/6N, 26-30g). Functional parameters of the left ventricle were derived with Segment (Medviso AB, Lund, Sweden). The results were tested for significance with a paired Student’s t-test.Results
Figure 2 shows image quality for the different reconstruction mechanisms. Figure 3 shows Bland-Altman plots of the functional values.The CS reconstruction is very close to the reference standard in all evaluated parameters (end-diastolic volume (EDV), end-systolic volume (ESV), stroke volume (SV) and ejection fraction (EF)). EDV is slightly overestimated and the ESV exhibits no bias, leading to an increased SV and EF, although none of the observed differences are significant (p=0.18 (EDV), p=0.87 (ESV), p= 0.41 (SV), p=0.67 (EF)).
The direct deep learning reconstruction yields slight underestimation of the ESV and very slight underestimation of the EDV, again leading to increased SV and EF. The differences are not significant (p=0.21 (EDV), p=0.05 (ESV), p= 0.25 (SV), p=0.05 (EF)).
The residual deep learning reconstruction shows significant underestimation of EDV (p=0.04) and ESV (p=0.005). The SV is marginally increased (p=0.55), leading to a significant overestimation of the EF (p=0.005).
Discussion
Compared to the sliding window approach, image quality is improved with both DL reconstruction methods. The still images show a high level of detail in the heart, papillary muscles can be appreciated in the DL-, but not in the CS reconstructed image in Figure 2. However, intensity modulations over the time series degrade video (or M-Mode) quality (referred to as "flickering” artifact by Hauptmann et. al.1, and attributed to the slice-by-slice approach). Although not crucial for evaluation of functional parameters, the CS reconstructed movies appear smoother.Underestimation of the ESV with the real-time sequence and all reconstruction techniques may be explained by the acquisition and reconstruction: The gated reference-standard divides the cardiac cycle into bins and averages over multiple cycles. In contrast, the sliding-window approach enables the reconstruction of images at almost any point in time, which could lead to a better “targeting” of end-diastole. This underestimation is larger in DL methods, as the CS reconstruction utilizes temporal regularization, which might align the very short end-systolic phase with adjacent cardiac phases, not corresponding to full contraction.
The underestimation of EDV in both DL methods is difficult to explain. The bias of the direct model seems to be dominated by one outlier, most likely caused by motion of the animal. With residual DL, the bias seems to be more systematical, but as it is considerably smaller, it could also be attributed to pure chance. SV and EF are almost identical to the reference standard for compressed sensing, making it a promising candidate in terms of reproducibility of functional values. Both, direct and residual DL methods, overestimate the SV and result in an EF increase of approximately 2 percentage points.
Conclusion
This work demonstrates the feasibility of using 2D CNNs for real-time imaging in rodent MRI. The results are promising and mostly in line with theoretical expectations. Further research into the amount and quality of training data and different network architectures is necessary to achieve a similar robustness to the established CS reconstruction method, e.g. reduction of flickering .A comparison to the gated reference standard with equal temporal and spatial resolution could be helpful in understanding the bias in the functional values.Acknowledgements
The authors thank the Ulm University Centre for Translational Imaging MoMAN for its support.References
[1] Hauptmann et. al., “Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning – proof of concept in congenital heart disease”, DOI: 10.1002/mrm.27480, Magn Reson Med. 2019,81:1143-1156
[2] Kofler et. al., “ Spatio-Temporal Deep Learning-Based Undersampling Artefact Reduction for 2D Radial Cine MRI with Limited Training Data”, IEEE:TMI
[3] Animal Care and Use Committee, Johns
Hopkins University (http://web.jhu.edu/animalcare/procedures/mouse.html#normative, retrieved 11/06/2019)
[4] Wundrak, Stefan, et al. "Golden ratio sparse MRI using tiny golden angles." Magnetic resonance in medicine 75.6 (2016): 2372-2378.
Figures
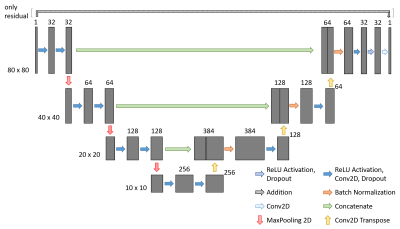
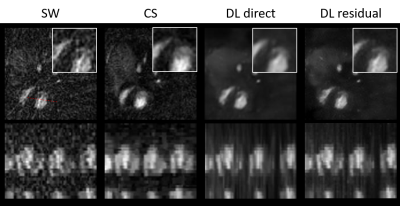
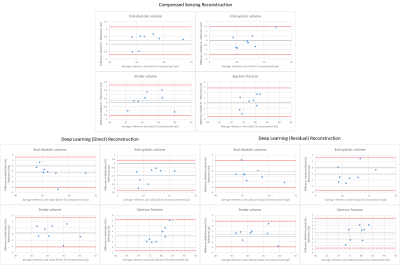
Figure 3: Bland-Altman plots for functional values derived from the CS (a), direct DL (b) and residual DL (c) reconstructed movies.
(a) The end-diastolic volume is slightly overestimated and the end-systolic volume exhibits no bias. This results in an increased stroke volume and almost identical ejection fraction.
(b) EDV and ESV are underestimated, SV very slightly overestimated. This leads to an increased EF.
(c) End-diastolic and end-systolic volumes are underestimated, the stroke volume is slightly overestimated, resulting in an increased ejection fraction.