2235
Reducing pulsatile artefact in TOF sequence using CNN1Central Research Institute, United Imaging Healthcare Group, United Imaging Healthcare, Shanghai, China, 2Magnetic Resonance Business Unit, United Imaging Healthcare, Shanghai, China, 3Department of Radiology, Peking University Third Hospital, Beijing, China
Synopsis
Time-of-flight (TOF) sequence is a non-contrast enhancing blood vessel imaging technique critical for vessel wall analysis. However, the periodical pulsation in the vessels, especially arteries, often causes pulsatile artifact along the phase encoding direction in the image. This degrades image quality and brings difficulty for diagnosis. The purpose of this work is to develop a convolutional neural network (CNN) method to reduce the pulsatile artefact in TOF sequence.
Introduction
Time-of-flight (TOF)1 sequence is commonly used for vessel imaging without the need for contrast. The periodical pulsation in the vessels, especially arteries, often causes pulsatile artifact along the phase encoding direction in the image. This degrades image quality and brings difficulty for diagnosis, especially for vessel wall analysis2. The purpose of this work is to develop a convolutional neural network (CNN) based solution to reduce the pulsatile artefact in TOF sequence.Methods and Materials
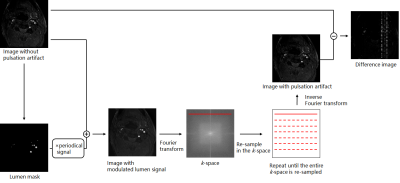
Data acquisition20 TOF series form MR scans (uMR 770, UIH, Shanghai, China) were used for paired training dataset generation, as shown in Figure 1. 1) a vessel mask in the original image without pulsation artifact was generated using an intensity threshold method. Then a periodically changing signal that was designed according to the ECG signal and sequence TR, was applied to the vessel mask to mimic the lumen signal modulation. 2) The image with lumen signal modulation was Fourier transformed into k-space, and re-sampled by one TR. 3) This process repeats until all the k-space lines are re-sampled. 4) After inverse Fourier transform, the re-sampled image contained the pulsation artifact. 5) Finally, a difference map image was obtained by subtracting the original image from the image with pulsation artifact.
Network architecture
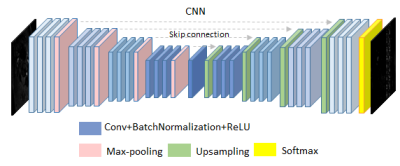
We used a 2D Unet with dense connections 4,5 in this work. It is made up of four encoders and four decoders connected by a bottleneck layer and skip connections linking every decoder with the encoder of the same spatial resolution. Each encoder or decoder consists of 3 convolution, batch normalization and RuLU layers respectively, with skip connections improving gradient flow. The final classification layer is a softmax-activated convolution layer with a kernel size of 1x1. We used mean squared error as loss function and Adam as optimizer.
Training
Out of 20 TOF series, we randomly choose 3 series as testing data and 2 series with noticeable pulsatile artifact as validation data. The input images were resized to 256 x 256 and normalized by z-score before entering the CNN. We trained the model twice with series randomly rotated 90 degrees for data augmentation.
Results
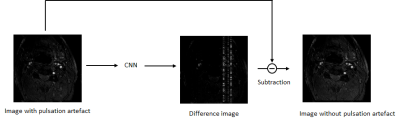
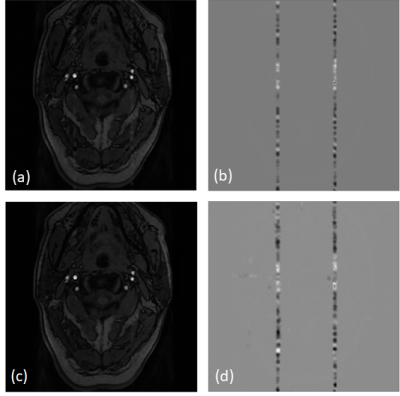
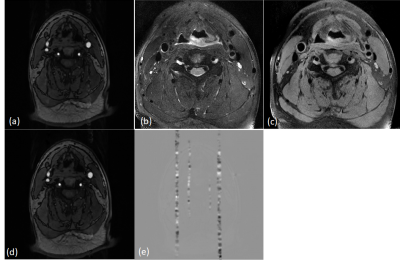
We evaluated our trained model on testing and validation data. Qualitatively, the predictions of testing data show great resemblance with ground truth images, where a spurious band of heterogeneous shadow (similar to zipper artifact) threads the transversal section of the artery, as shown in Figure 4. Quantitatively, we calculated the structural similarity index (SSIM) of recovered images and original artifact-free image. The SSIM was 0.928±0.024. In our validation data, our model could effectively extract the band-like artifact from the original image. By subtraction the prediction from the original image, the disturbance of pulsatile was removed, as shown in Figure 5.Discussion
Pulsate artifact in TOF series are caused by periodic spatial movement of the blood in vessels that disturb phase encoding, and would affect clinical diagnosis. Herein we propose a method to artificially generate this kind of artifact, which can be used for deep learning. To remove the artifact, we simply subtracted the predicted difference map from the image with artifact. While predictions of generated images showed great resemblance to the ground truth images and could effectively remove the artifact, some of the real images had too severe artifacts that were hard to remove completely. This is possibly because these severe artifacts were caused by more complex physical movements that exceeds the scope of our simulation. We believe that with further validation and more training cases, this approach will become more robust and have great clinical value.Conclusion
In this work we have proposed a deep learning solution for pulsatile artifact reduction. We prepared paired training images by resampling in k-space that mimics phase encoding failure caused by blood flow. We trained a Unet that extracts pulsatile artifact from TOF series and evaluated the model on both artificial and real-life artifact-infected images. Both SSIM and visual interpretation show that our model could effectively extract pulsate artifact from TOF images and thus improve image quality.Acknowledgements
No acknowledgement found.References
1. Ota H, Yarnykh VL, Ferguson MS, Underhill HR, Demarco JK, Zhu DC, Oikawa M, Dong L, Zhao X, Collar A, Hatsukami TS, Yuan C. Carotid intraplaque hemorrhage imaging at 3. 0-T MR imaging: comparison of the diagnostic performance of three T1-weighted sequences. Radiology. 2010;254(2):551–563.
2. Perman W H , Moran P R , Moran R A , et al. Artifacts from pulsatile flow in MR imaging[J]. J Comput Assist Tomogr, 1986, 10(3):473-483.
3. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation[J]. 2015
4. Roy A G , Conjeti S , Navab N , et al. QuickNAT: A Fully Convolutional Network for Quick and Accurate Segmentation of Neuroanatomy[J]. 2018.
Figures